A clustered regularly interspaced short palindromic repeats knockout method to reveal methyl-CpG binding domain 4 function
Received: 14-Dec-2022, Manuscript No. PULJMBR-22- 5894; Editor assigned: 16-Dec-2022, Pre QC No. PULJMBR-22- 5894 (PQ); Accepted Date: Jan 05, 2023; Reviewed: 24-Dec-2022 QC No. PULJMBR-22- 5894 (Q); Revised: 29-Dec-2022, Manuscript No. PULJMBR-22- 5894 (R); Published: 10-Jan-2023, DOI: 10.37532/puljmbr.2023.6(1).1-4
Citation: Ng AH. A clustered regularly interspaced short palindromic repeats knockout method to reveal methyl-CPG binding domain 4 function. J Mic Bio Rep. 2023;6(1):1-4.
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
DNA methylation is an epigenetic mechanism tailored for DNA repression, engineered for regulating gene expression without direct manipulation of the nucleotide sequence. One component of this process includes Methyl-Binding Proteins (MBD), which have an affinity for methyl groups, and they competitively inhibit transcription factors from binding with genetic promoters. Interestingly, MBD4 is unique because, as opposed to transcriptional repression, it promotes gene repair & demethylation and is associated with various methylation-related diseases, such as Autism. By further studying MBD4, we can identify a potential therapeutic target for MRD and further understand the role of methylation on the epigenome regarding seasonal plasticity. Therefore, this paper describes a CRISPR Knockout screen to isolate & repress MBD4 from its customary functionality with optimized gRNA targets in Astatotilapia burtoni Cichlid. I expect a morphological change in the Cichlid’s skin color (such change can be identified with computer vision COCO-StyleDataset-Generator-GUI), which substantiates my belief that MBD4 does play a significant role in seasonally-regulated epigenetic switches and can be targeted in methylation treatments. However, the exogenous factors relating to MBD4’s role in methylation remain to be investigated.
Keywords
Epigenetics; MBD4; Methylation; Astatotilapia burtoni; CRISPR; Plasticity
Introduction
Methylation is an epigenetic mechanism tailored for DNA repression, capable of regulating gene expression without directly editing the nucleotide sequence [1]. It functions by transferring a Methyl Group (CH3 ) from S-Adenyl Methionine (SAM) to the 5’ end of a cytosine (mC), via DNA Methyltransferases (DNMTs), especially within CpG dinucleotides [2]. This attracts a group of proteins that possess a particularly significant affinity for methyl groups, also known as Methyl-Binding Proteins (MBD). The majority of MBD domain proteins repress gene expression by binding to promoters, thus competitively preventing essential transcription factors from properly functioning [1]. They are also responsible for interacting with a host of other proteins, including histone deacetylases and chromatin, which disturb histone and heterochromatin structures, respectively [3,4]. Interestingly, one version of MBD demonstrates characteristics apart from repression, as MBD4 is evidenced to contribute towards gene repair in regards to CpG-TpG (methylated cases) and CpG-UpG (unmethylated cases) mismatches; the associated binding partners include MLH1, DNMT1- UHRF/USP7, and DNMT3B [5]. In addition to the N-terminal MBD domain that all MBD proteins possess, MBD4 also has a unique Cterminal glycosylate domain that catalyzes their DNA repair & demethylation characteristics [6]. There is evidence to suggest that MBD4 mutations/misregulation contribute to autism spectrum disorder, cancerous tumors (especially in the gastrointestinal tract), hematopoiesis illness, and even memory bias in injured mice with olfactory methylation [7-10]. Therefore, it is essential that we fully grasp the mechanisms behind MBD4 and its contribution to disease progression and ecology, which are both factors not yet well understood. For example, what role does MBD4 play in DNA methylation & disease? In what ways does MBD4 facilitate molecular plasticity in response to the environment? Does methylation contribute to seasonally transcriptional phenotypes or vice versa? [11].
As a result, targeting MBD4 in a CRISPR/Cas9 knockout screen would reveal the protein function necessary for further understanding the role of methylation in the human body, especially regarding the pathogenesis of the disease. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) is a genetic-editing tool that is used to alter sequences of DNA. Cas9 cleaves DNA, which allows for the insertion or removal of the desired gene, and it targets that gene with a sgRNA binding sequence matching that of the desired target sequence [12]. In a CRISPR knockout screen, CRISPR/ Cas9 would cleave the target site, which would generate random misplaced insertions/deletions (indels) as a result of non-homologous end joining. These “mistakes” from NHEJ DNA repair pave the way for (frameshift) mutations, thus rendering the gene dysfunctional to perform reverse genetics [13]. In other words, knockouts include inhibiting gene expression and observing the phenotypic consequences to infer gene function. The CRISPR knockout screen protocol can also be slightly modified for fish subjects [14]:
• Breed wild-type fish and collect the founder eggs.
• Synthesize sgRNAs for high indel rates.
• Microinject the sgRNA with CRISPR/Cas9 into founder eggs.
• Phenotype the resulting founding adult Cichlid (F0 generation).
• Outcross the mosaic F0 adults with wild types.
• Cross the confirmed F1 heterozygotes and sequence for mutations.
• Phenotype the homozygous F2 generation.
Astatotilapia burtoni fish (blue-yellow color morph) serves as the ideal model for a knockout screen, given the reliability of their observable plastic phenotypes. For instance, such males exhibit dominant sexual behavior and are known to be aggressive when protecting their territory during mating season. Though, this behavior can be easily influenced by its dynamic social environment, making A. burtoni a reliable model to study the mechanisms in which methylation could contribute to their plastic behavioral traits [15]. The upregulation of various neuroendocrine genes in territorial males suggests that the “socially-regulated phenotype switch” is particularly apparent in this species of Cichlid, and observable color changes in Cichlid skin have similarly been exploited for reverse genetics [16,17]. Therefore, the unique plastic phenotype of A. burtoni allows us to study the effect of the environment on epigenetic genomics and will be the model of choice as a result.
I hypothesize that MBD4 plays an important role in the environmentallyregulated switch that Cichlid phenotypes (behaviorally & physiologically) function, especially given the host of evidence demonstrating a correlation between methylation levels and transcriptional phenotypes [11]. For instance,Hibernation-Specific Protein-27 (HP-27) in chipmunks is upregulated during hibernation, suggesting a seasonal or situational effect on epigenetics [18]. Since MBD4 is directly associated with how methylation functions, it’s reasonable to infer that MBD4 would play a similar role. Of course, there is limited evidence to suggest that MBD4 directly affects the adaptive plasticity of vertebrate species (as opposed to exogenous factors), which is the main factor to be investigated in this study [3,4]. Therefore, if knocking out the MBD4 gene in A. burtoni fish presents any significant alterations in phenotype, it can be assumed that MBD4 is the main contributor to plasticity or the earliest contributor to phenotype in the methylation pathway. This could reveal a therapeutic target for methylation-related illnesses and hint at the role of epigenetics on the genome in a dynamic environment [7-10].
Materials and Methods
Subjects
I used Astatotilapia burtoni fish, including 20 females/1 male, collected from Lake Malawi [19]. Adult fish will be held in 30 L tanks with regulation pH (8.0-8.2), salinity (320 ppm-480 ppm), and temperature (25°C) with 12-hour light-dark schedules. All experimental procedures will be approved by the Animal Care Committee [20].
In mating, all fish will be injected with Ova prim (Syndel) at 0.5 μL per gram of body mass and allowed to mate for 30 minutes. Embryos (flushed from the oral cavity) will be collected with 4mm transfer pipettes and stored in well plates treated with 6 mL of 1mg/L methylene blue antifungal reagent (Sigma) water after collection. 100 embryos will be used (80 experimental, 20 negative control) since the evidence shows that it takes 6 founder fish to chance a mutation [21].
Experimental procedure overview
I performed a CRISPR/Cas9 knockout screen on the MBD4 gene. This includes a CRISPR/Cas9 injection into A. burtoni embryos to breed the founder F0 generation. I then will outcross the founder and wild-type fish to limit off-targets, and will later screen the F1 generation for mutation germline transmission with fluorescent PCR & sequencing. Lastly, after crossing the mutant heterozygotes, the homozygous F2 generation will be finally examined for a phenotype. Untreated wild-type genotypes & phenotypes are used as controls. The procedures below follow the workflow described here in this overview.
sgRNA design and synthesis & cas9 preparation
I summarize below the set of rules that have been previously established for effective gRNA design along with my experimental specifics in parentheses immediately following each rule [22-24]:
• The target gene must be defined (MBD4)
• Cas9 protein & PAM sequence must be defined (SpCas9 & NGG)
• gRNA Promoter must be defined for expression (T7 Polymerase)
• High GC Content (45%-90%), limited predicted off-targets (<500), and exon targeting (close to 5’ end; region should be present in all isoforms and a functional protein domain) are also important factors that were considered
I performed a CRISPR/Cas9 knockout screen on the MBD4 gene. This includes a CRISPR/Cas9 injection into A. burtoni embryos to breed the founder F0 generation. I then will outcross the founder and wild-type fish to limit off-targets, and will later screen the F1 generation for mutation germline transmission with fluorescent PCR & sequencing. Lastly, after crossing the mutant heterozygotes, the homozygous F2 generation will be finally examined for a phenotype. Untreated wild-type genotypes & phenotypes are used as controls. The procedures below follow the workflow described here in this overview.
Remote grna sequences can be found in the results
The target sequences were then cross-checked with Synthego’s Guide Design Verifier to confirm the effectiveness of each sequence as suggested by CHOPCHOP. The final oligonucleotides will be created by annealing together a 5’ T7 promoter and a 3’ overlapping gRNA sequence to the target sequence, not including the PAM site (ordered on IDT). This is considered the top strand:
5’ - TAATACGACTCACTAT - gRNA - GTTTTAGAGCTAGAAATAGC - 3’
Remote full oligos can be found in results
To synthesize the crRNA-tracRNA, I will use a recently developed cloningfree method that requires the expansion of the top strand and a universal bottom strand: (AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTA GCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC) [25]. I will mix the top & bottom strand solutions and use PCR amplification to extend the new oligo to a confirmed 120 bp according to protocol. Lastly, I will combine a T7 RNA synthesis mix to the new oligo, PCR the solution, add DNAse, and purify the final reaction, all according to the manufacturer’s instructions. The resulting 2 sgRNAs (2 targets) will be stored at -80°C and will be co-injected.
Cas9 mRNA will be derived by digesting pT3TS-NLS-zCas9-NLS plasmid with XbaI and preparing RNA transcription reaction with message machine T3 kit (Life Technologies) [26].
Microinjection
I prepared micropipette needles according to methods used in Sticklebacks with MPPI-3 Pressure Injector (Applied Scientific Instrumentation), adopting air supply, pressure, dissecting microscope magnification, and other accepted settings [27]. The needles will be held on ice vertically in a capillary storage jar.
To prepare the microinjection contents, I will combine 200 ng of the synthesized sgRNA (25 pg), 400 ng of Cas9 mRNA (300 pg), 0.5μl of 0.5% DPBS (Elabscience, EP-CM-L0409), and 5μl of RNAse free water. I will then pipet these contents into the blunt end of the needle at 0.5 μl increments until the needle is filled and the very end of the needle is broken with the watchmaker’s forceps at a 60° angle. 25 minutes’ post-fertilization, I will separate the embryos into each indentation of a saw blade and flush each embryo with water for 2 seconds to swell the chorion. To start the injection, I will rotate the embryos until the blastomere is directly perpendicular to the injection needle, and then push the needle into the cytoplasm outside of the yolk. I inject the solution with a foot pedal until a red spot with diffuse edges fills about one-eight of the cytoplasm’s diameter. A transfer pipette will be used to transfer the injected embryos into the well plates. I will check the health of these embryos daily.
Verify mutations with fluorescent pcr & sequencing
At 48 hours’ post-injection, I will collect diluted DNA samples from the Cichlid embryos with Extract-N-Amp (Sigma, E7526), according to the manufacturer’s instructions. Additionally, I designed primers to begin fluorescent PCR by adding an M13F sequence to the forward primer and a PIG-tail sequence to the reverse primer, all annealed at the 5’ end of the gene-specific primers suggested by CHOPCHOP.
Remote designed primers can be found in the results
I will create the fluorescent primer mix by mixing 5 μl of this 100 μM forward/reverse primers, 485 μl of TE pH 8.0 (Quality Biological, cat no. 351-011-131), and 5 μl of 100 μM M13-FAM primer (/56-FAM/ TGTAAAACGACGGCCAGT). I will then add 6 μl of the fluorescent primer mix to 100 μl of PCR master mix to synthesize the final mix, and finally, add 1.5 μl of diluted DNA to 5 μl of the final mix. The resulting solution is deemed the analyzing product [21].
To start fluorescent PCR, I will amplify the analyzing product in a thermocycler following these conditions: 94°C, 12 min, denature cycle 1; 94°C, 30 seconds, denature cycle 2-36; 55°C, 30 seconds, anneal cycle 2-36; 72°C, 30 seconds, extend cycle 2-36; 72°C, 10 minutes, extend cycle 37; 4°C, hold cycle 38. I will then analyze the resulting product with a Genetic Analyzer 3130xl on CRISPR-STAT parameters [28]. GeneMapper with the accepted settings in combination with Peak Scanner 2 (Thermofisher) can be used to display peak sizes from 3130xl, and the embryos that illustrate frameshift mutations, shown by more than 1 peak present, will be incubated to start the mutant lines [28]. A low ratio between areas under the wild-type peak vs all peaks indicates high mutation frequency [14]. In adult Cichlid, I will examine DNA from their servered caudal fin. Sanger Sequencing with 3730xl and PolyPeakParser provides the exact nucleotide sequence to confirm genotype and allele mutation [29]. I will lastly examine the fish for phenotypic expressions as a result of the induced mutation. Observations can be identified automatically with computer vision COCO-Style-DatasetGenerator-GUI.
Results
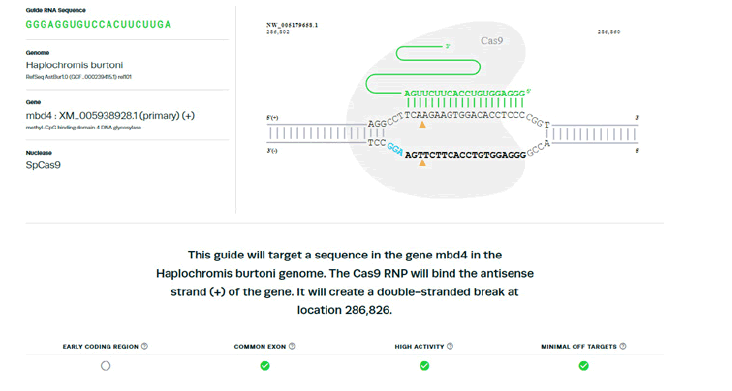
Presented are the computational results for designer strands:
gRNA
5’ - GGGAGGTGTCCACTTCTTGAAGG - 3’
5’ - GGAAGGGGGTGCTTGTGATGTGG - 3’
These sequences were verified for efficacy with Synthego software. (Figure 1).
Figure 1: Synthego verification. Target sequence GGGAGGTGTCCACTTCTTGAAGG (PAM GGA) correlates with gRNA sequence GGAGGUGUCCACUUCUUGA (Above). There’s evidence that this gRNA will target a common exon, exhibit high activity, and have minimal off-targets. Target sequence GGAAGGGGGTGCTTGTGATGTGG (PAM GGT) correlates with gRNA sequence GGAAGGGGGUGCUUGUGAUG (Below). There’s evidence that this gRNA will target a common exon in an ear
Oligos
Created by annealing a T7 promoter and an overlapping universal sequence to the gRNA targets, as explained in the methods section
5’-TAATACGACTCACTATGGGAGGTGTCCACTTCTTGAGTTTTAGAGCTAGAAATAGC–3’
5’-TAATACGACTCACTATGGAAGGGGGTGCTTGTGATGGTTTTAGAGCTAGAAATAGC–3’
Primer
Target: GGGAGGTGTCCACTTCTTGAAGG
Forward:
5’- TGTAAAACGACGGCCAGTCCTTATTTCAGTGGGAAATCCA - 3’
Reverse:
5’ - GTGTCTTCAGGGATCATGGAAAAGAGTTT - 3’
Target: GGAAGGGGGTGCTTGTGATGTGG
Forward:
5’- TGTAAAACGACGGCCAGTTTCACCACAGAAGCCCTTAGAT - 3’
Reverse:
5’-GTGTCTTGAGGTGCCTCTAGAATTGCTGT-3’ly coding region, exhibit high activity, and have minimal off-targets (Figure 2)
Figure 2: Primer synthesis. Target GGGAGGTGTCCACTTCTTGAAGG is calculated to have left primer CCTTATTTCAGTGGGAAATCCA and right primer CAGGGATCATGGAAAAG7jAGTTT (3). Target GGAAGGGGGTGCTTGTGATGTGG is calculated to have left primer TTCACCACAGAAGCCCTTAGAT and right primer GAGGTGCCTCTAGAATTGCTGT (4). Efficiency is centered around 60.0, and base pair length is centered around 245 bp for each primer
Discussion
Given the demonstrated efficiency of the designer strands, the probability of success is significantly high in this experiment. gRNA strands have been optimized for efficiency. Between my selected target strands GGGAGGTGTCCACTTCTTGAAGG and GGAAGGGGGTGCTTGTGATGTGG, both exhibited satisfactory qualities for a traditional CRISPR knockout, including a high GC content, limited off-targets, and wide exon targeting [21]. Primers were also optimized for a CRISPR knockout, especially regarding base pair length from 200bp300 bp [24]. After injecting the CRISPR knockout solution, we can observe phenotype either manually or with COCO-Style-Dataset-Generator-GUI, as described earlier.
Conclusion
Therefore, I conclude that this CRISPR KO experiment will function effectively and will reveal that the MBD4 gene plays a significant role in an environment-regulated switch that facilitates phenotypic plasticity. CRISPR knockout will likely disable MBD4 gene expression theorized to play a key role in DNA methylation. Additionally, it’s well known that HibernationSpecific Protein-27 (HP-27) in chipmunks is upregulated during hibernation, suggesting seasonal regulation on epigenetics. By relation, it’s inferrable that DNA methylation, an epigenetic process, is similarly manipulated via MBD4 with seasons. In Cichlids, this relationship can usually be indicated by a morphological physical change MBD4 that would result in a difference in skin color and/or sexual behavior in comparison to the control MBD4- intact Cichlid. This is particularly significant because the unique plastic phenotype of A. burtoni allows us to study the effect of the environment on epigenetic genomics and provides us the opportunity to reveal a direct correlation between MBD4 & plasticity. A change in skin color and/or behavior is evidence of such a proposition and could reveal a therapeutic target for methylation-related illnesses.
However, many questions are still left unanswered: What exogenous factors could still affect the phenotypic expressions of methylation? What characteristics of each season prompt molecular plasticity? How can we apply the role of MBD4 on methylation in treatments.
References
- Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23-38.
- Albalat R, Marti-Solans J, Canestro C. DNA methylation in amphioxus: from ancestral functions to new roles in vertebrates. Briefings in functional genomics. 2012;11(2):142-55.
- Ng HH, Zhang Y, Hendrich B, et al. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nature genetics. 1999 ;23(1):58-61.
- Santoro R, Grummt I. Epigenetic mechanism of rRNA gene silencing: temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Molecular and cellular biology. 2005;25(7):2539-46.
- Du Q, Luu PL, Stirzaker C, et al. Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics. 2015 ;7(6) :1051-73.
- Manvilla BA, Maiti A, Begley MC, et al. Crystal structure of human methyl-binding domain IV glycosylase bound to abasic DNA. Journal of molecular biology. 2012 ;420(3):164-75.
- Cukier HN, Rabionet R, Konidari I, et al. Novel variants identified in methyl-CpG-binding domain genes in autistic individuals. Neurogenetics. 2010;11(3):291-303.
- Wong E, Yang K, Kuraguchi M, et al. Mbd4 inactivation increases C→ T transition mutations and promotes gastrointestinal tumor formation. Proceedings of the National Academy of Sciences. 2002;99(23):14937-42.
- Chen ZP, Gu DS, Zhou ZP, et al. Decreased expression of MBD2 and MBD4 gene and genomic-wide hypomethylation in patients with primary immune thrombocytopenia. Human Immunology. 2011 ;72(6):486-91.
- Tajerian M, Alvarado SG, Clark JD. Differential olfactory bulb methylation and hydroxymethylation are linked to odor location memory bias in injured mice. Molecular Pain. 2019; 15:1744806919873475.
- Alvarado S, Fernald RD, Storey KB, et al. The dynamic nature of DNA methylation: a role in response to social and seasonal variation. Integrative and comparative biology. 2014 ;54(1):68-76.
- Rodriguez‑Rodriguez DR, Ramírez‑Solís R, Garza‑Elizondo MA, et al. Genome editing: A perspective on the application of CRISPR/Cas9 to study human diseases. International journal of molecular medicine. 2019;43(4):1559-74.
- Bock C, Datlinger P, Chardon F, et al. High-content CRISPR screening. Nature Reviews Methods Primers. 2022 ;2(1):1-23.
- Li CY, Steighner JR, Sweatt G, et al. Manipulation of the Tyrosinase gene permits improved CRISPR/Cas editing and neural imaging in cichlid fish. Scientific Reports. 2021;11(1):1-2.
- Fernald RD, Hirata NR. Field study of Haplochromis burtoni: habitats and co-habitant. Environmental Biology of Fishes. 1977;2(3):299-308.
- Renn SC, Carleton JB, Magee H, et al. Maternal care and altered social phenotype in a recently collected stock of Astatotilapia burtoni cichlid fish. Integrative and comparative biology. 2009 ;49(6):660-73.
- Alvarado SG. Molecular plasticity in animal pigmentation: emerging processes underlying color changes. Integrative and Comparative Biology. 2020;60(6):1531-43.
- Fujii G, Nakamura Y, Tsukamoto D, et al. CpG methylation at the USF-binding site is important for the liver-specific transcription of the chipmunk HP-27 gene. Biochemical Journal. 2006;395(1):203-9.
- Malinsky M, Svardal H, Tyers AM, et al. Whole-genome sequences of Malawi cichlids reveal multiple radiations interconnected by gene flow. Nature ecology & evolution. 2018;2(12):1940-55.
- Fernald RD. Quantitative behavioural observations of Haplochromis burtoni under semi-natural conditions. Animal Behaviour. 1977; 25:643-53.
- Varshney GK, Pei W, LaFave MC, et al. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome research. 2015;25(7):1030-42.
- Mohr SE, Hu Y, Ewen‐Campen B, et al. CRISPR guide RNA design for research applications. The FEBS journal. 2016 ;283(17):3232-8.
- Shi J, Wang E, Milazzo JP, et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nature biotechnology. 2015;33(6):661-7.
- Varshney GK, Carrington B, Pei W, et al. A high-throughput functional genomics workflow based on CRISPR/Cas9-mediated targeted mutagenesis in zebrafish. Nature protocols. 2016;11(12):2357-75.
- Montague TG, Cruz JM, Gagnon JA, et al. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic acids research. 2014 ;42(W1): W401-7.
- Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proceedings of the National Academy of Sciences. 2013;110(34):13904-9.
- Erickson PA, Ellis NA, Miller CT. Microinjection for transgenesis and genome editing in threespine sticklebacks. JoVE (Journal of Visualized Experiments). 2016; 13(111): e54055.
- Carrington B, Varshney GK, Burgess SM, et al. CRISPR-STAT: an easy and reliable PCR-based method to evaluate target-specific sgRNA activity. Nucleic acids research. 2015 ;43(22):e157.
- Hill JT, Demarest BL, Bisgrove BW, et al. Poly peak parser: Method and software for identification of unknown indels using sanger sequencing of polymerase chain reaction products. Developmental Dynamics. 2014 ;243(12):1632-6.