Cellular uptake and cytotoxicity studies of pH-responsive polymeric nanoparticles fabricated by dispersion polymerization
Received: 06-Mar-2018 Accepted Date: Apr 02, 2018; Published: 12-Apr-2018
Citation: Puri R, Adesina S, Akala E. Cellular uptake and cytotoxicity studies of pH-responsive polymeric nanoparticles fabricated by dispersion polymerization. J Nanosci Nanomed. April-2018;2(1):3-16.
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
Objective: A strategy in site-specific drug delivery is the use of pHgradients that exist in diseased conditions such as cancer for the release of loaded drug(s) in the biophase. The objective of this work is to synthesize pH-responsive docetaxel-loaded nanoparticles with a bisacrylate acetal crosslinker, which can get internalized into cells, and which will be equivalent to or more cytotoxic than the free drug against cancer cells. Methods: pH-responsive nanoparticles were synthesized by a dispersion polymerization technique. The nanoparticles were characterized for physicochemical properties. Cytotoxicity studies of the nanoparticles were performed on PC3 and LNCaP prostate cancer cell lines using a cell viability assay. Cellular uptake studies were performed using a confocal laser scanning microscope. Results: Smooth spherical nanoparticles were formed. In-vitro drug release was faster at pH 5.0 than pH 7.4, which confirmed the pHresponsiveness of the nanoparticles. Cytotoxicity studies showed that the nanoparticles were more effective at the same molar amount than the free drug against cancer cells. Both dose exposure and incubation time affected the cytotoxicity of prostate cancer cells. Furthermore, LNCaP cells appeared to be the more sensitive to docetaxel than PC3 cells. The cellular uptake studies clearly showed the presence of discrete nanoparticles within the cells in as little as 2 hours. Conclusion: pH-sensitive nanoparticles were developed; they degraded quickly in the mildly acidic environments similar to those found in endosomes and lysosomes of tumor tissues. These novel pH-sensitive nanoparticles would offer several advantages over conventional drug therapies.
Keywords
Acetal, crosslinker, pH-responsive, dispersion polymerization, polymeric nanoparticles, hydrolysis, cell viability assay, cytotoxicity and cellular uptake
Introduction
Prostate cancer (PCa) is the most common solid organ malignancy affecting men and the second leading cause of cancer death, following lung cancer in men in the United States [1]. Docetaxel is one of the most effective anticancer drugs for the treatment of prostate cancer [2,3]. However, the commercial formulation of docetaxel (Taxotere®) is presented as a solution with a high amount of solubilizers (Tween 80/ ethanol). A high concentration of these solubilizers in its formula has been associated with severe toxic effects and allergic reactions. Hence, it is necessary to develop alternative formulations without the use of these solubilizers to prevent the side effects and also to target docetaxel specifically to tumors cells [4,5]. Nanotechnology platform can be used for site-specific drug delivery of docetaxel to the cancerous cells. Nanoparticles, by using the passive targeting (enhanced permeability and retention effect-EPR) and active targeting (by ligand-receptor or antigenantibody interactions and receptor mediated endocytosis) can result in increased accumulation and internalization or intracellular uptake of the nanocarriers into the tumors. However, these strategies have some limitations [6-8]. The EPR effect can enhance the accumulation of nanoparticles in tumor tissues but it may not result in good cellular internalization as well as sufficient intracellular drug release [6].
Current approaches for the development of pH-sensitive nanocarrier systems generally involve either incorporation of “titratable” groups into the copolymer or the use of linkages (crosslinkers) that degrade under pH changes. Several pH-sensitive linkages have been proposed in recent years for the development of nanoparticles and polymer-drug or antibody-drug conjugates. pH-sensitive covalent bonds such as hydrazone, orthoester, cis-aconityl, N, O-dimethacryloylhydroxylamine and acetal are used as linkages to develop acid-labile nanoparticles [9-11]. This linkage can be used either to directly attach the drug to the copolymer or to alter the structure of the polymer sufficiently that drug release is triggered upon hydrolysis of the linker [12].
An ideal acid-sensitive linker in a drug delivery system would have an increased rate of hydrolysis at pH~5.5, but be fairly stable at physiological conditions (pH=7.4). Though various environmental responsive stimuli are used in drug delivery, pH-responsiveness is the most frequently used, as pH in the tumor microenvironment is slightly more acidic than in the normal cells. In addition, pH-responsiveness has been proposed for nanocarriers taken up by cells via an endocytosis process [13]. Although the endocytic pathway of cells begins near the physiological pH of 7.4, it drops to a lower pH of 5.5-6 in the endosomes and approaches pH 5.5-5 in the lysosomes. Therefore, nanocarriers that are responsive to these gradients can be designed to release their payload selectively within tumor cells [+].
In this work, free radical dispersion polymerization technique was used for the fabrication of pH-sensitive nanoparticles. N-butyl acrylate (n-BA) was copolymerized with the acid cleavable bisacrylate acetal crosslinker. Poly (ethylene glycol) was used as a steric stabilizer which, when attached onto the surface of the nanoparticles, rendered the surface hydrophilic (stealth nanoparticles). Stealth nanoparticles are not recognized by the reticuloendothelial system and hence have increased circulation time and accumulation into the tumors by enhanced permeability and retention effect. Also, once the nanoparticles accumulate into the tumor environment, get internalized into cancerous cells by endocytosis and then reach the acidic organelles such as endosomes (pH 5.5-6) and lysosomes (pH 5-5.5), the acetal crosslinker in the nanoparticles hydrolyzes, resulting in the disruption of nanoparticle structure and the release of loaded docetaxel into the cells, resulting in site-specific drug delivery.
Benzaldehyde acetals are well-suited for the preparation of acid-labile nanoparticles because their acid-sensitivity can be tuned by the introduction of substituents at different positions on the aromatic ring [14]. The use of crosslinkers containing substituted benzaldehyde acetals has been shown to provide an easy and versatile way to introduce acid cleavability to polymeric colloidal systems for site-specific drug release [15,16]. Akala and his coworkers have synthesized three benzaldehyde bisacrylate acetal crosslinkers using three benzaldehydes: 4- methoxybenzaldehyde, 2,4- dimethoxybenzaldehyde and 2,4,6- trimethoxybenzaldehde [17]. The three crosslikers differ in the number of methoxy groups that are present on the aromatic ring. N-butyl acrylate monomer and the acetal crosslinkers were used for in situ nanoparticle preparation using BPO/N-PDEA as the redox co-initiator system. Poly (ethylene glycol) (n) monomethyl ether monomethacrylate (MW of PEG Block=1,000) was used both as a hydrophilic macromonomer and a steric stabilizer in the nanoparticle preparation. The results for the hydrolysis studies of the pure crosslinkers and nanoparticles containing the crosslinkers revealed that the rate of hydrolysis depended on the structure of the crosslinkers; it was also highly pH dependent. The rate of hydrolysis was faster at pH 5.0 buffer than pH 7.4 buffer. The nanoparticles formulated with di(2-methacryloyloxyethoxy)-[2,4,6- trimethoxyphenyl]methane crosslinker showed the fastest hydrolysis rate, followed by di(2-methacryloyloxyethoxy)-[2,4-dimethoxyphenyl]methane crosslinker and di(2-methacryloyloxyethoxy)-[4-methoxyphenyl]methane crosslinker. The data were similar to the results obtained for the hydrolysis of pure crosslinkers. In these structure activity relationship studies, the rate of hydrolysis of the crosslinker which has three methoxy groups was the fastest, followed by the crosslinker with two methoxy groups and then the crosslinker with one methoxy group on the benzaldehyde ring. The belief is that protonation of the acetal yields a resonance stabilized carbocation, which facilitates the hydrolysis of the crosslinkers to form the relevant alcohol and the aldehyde [17]. The acceleration of the kinetics of hydrolysis of the nanoparticles containing the crosslinkers at pH 5.5 buffer compared to pH 7.5 buffer is expected because the rate of hydrolysis of benzaldehyde acetals is proportional to the hydronium ion concentration [14,18].
Akala and his coworkers [17,19-24] have reported on in situ dispersion polymerization for the fabrication of nanoparticles. The polymerization technique involves the use of a macromonomer or a monomer that forms the core of the nanoparticles, a cross-linker that cross-links the core, a redox co-initiator system and a hydrophilic macromonomer (polyethylene glycol [PEG]), which confers stealth property to the nanoparticles and which also serves as a stabilizer. The advantages of nanoparticles fabrication by dispersion polymerization are as follows: fabrication of the nanoparticles is a one-pot process (simultaneous encapsulation of drugs during fabrication of nanoparticles by dispersion polymerization) carried out at an ambient temperature suitable for product development of thermo labile bioactive agents. It is possible to add surface functionalities in onebatch process without further modifications compared to nanoparticle fabrication by dispersion of preformed polymers which involves many steps leading to drug loss from the nanoparticles. The process is surfactant free thereby obviating problems associated with the use of surfactants [17,22].
Material and Methods
Materials
2,4-dimethoxybenzaldehyde and p-toluene sulfonic acid monohydrate were purchased from Sigma Aldrich (St. Louis, MO, USA). 2- hydroxyethyl methacrylate (HEMA) (97%) and n-butyl acrylate (99%) obtained from Sigma Aldrich were dried over molecular sieves (4A°) for 24 hours and distilled under negative pressure prior to use. Silica gel was purchased from Selecto Scientific Inc., USA. Molecular sieves (4A°), sodium hydroxide pellets, potassium phosphate monobasic, anhydrous dichloromethane (≥ 99.8%), triethylamine, ethyl acetate, n-hexanes, dichloromethane, methanol, sodium chloride, sodium acetate, potassium hydroxide, chloroform-D and docetaxel (purum, ≥ 99%) were obtained from Sigma Aldrich. Poly (ethylene glycol) n monomethyl ether mono methacrylate (MW of PEG Block=1,000) was obtained from Polysciences, Inc. (Warrignton, PA, USA). Benzoyl peroxide (BPO) and N-phenyldiethanolamine (N-PDEA) were obtained from Sigma Aldrich. All the chemicals were used as received, unless specified. Gibco® RPMI media 1640, heat-inactivated fetal bovine serum (FBS), trypsin-EDTA, penicillin-streptomycin antibiotic (10,000 U/ml), Hanks' balanced salt solution (HBSS buffer), Hoechst® 33342 dye, CellMask™ orange plasma membrane stain and rhodamine-123 were obtained from Life Technologies, USA. Dimethyl sulfoxide-Hybri Max™ was obtained from Sigma Aldrich. 96 well, flat, clear bottom white polystyrene TC-treated microplates for cell viability studies were obtained from Corning Inc., USA. Glass bottom microwell dishes (35 mm-dish diameter) for cell uptake studies were obtained from MatTek Corporation, USA. PC3 cells (ATCC® CRL-1435™) and LNCaP cells (ATCC® CRL-1740™) were obtained from the American Type Culture Collection (ATCC, Manassas, VA). The cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum and 1% antibiotic (penicillin/streptomycin, v/v), and incubated at 37°C in a humidified environment of 5% CO2. The cells were passaged once a week and medium was changed twice a week. The human LNCaP and PC3 cell lines served as androgen sensitive and androgen insensitive human prostate cancer cell lines respectively.
Instrumentation
1H NMR and 13C NMR spectra were recorded on a Bruker AVANCE 400MHz NMR spectrophotometer. Reported chemical shifts (ppm) are relative to residual CDCl3 peak and coupling constants are reported in Hz. 1H NMR spectra were recorded at 400MHz while 13C spectra were recorded at 100MHz. The molecular weight of the crosslinker was determined by high resolution mass spectroscopy using thermo LTQ orbitrap mass spectrometer (MS) with Nano Electro Spray (NSI) technique. FT-IR spectrophotometric analysis of crosslinker was carried out using a Perkin Elmer Spectrum 100FT-IR spectrometer enabled with attenuated total reflectance (ATR) technology. The average particle size and zeta potential were determined by dynamic light scattering using a Zetasizer Nano-ZS (Malvern Instruments, USA). The mean of three measurements was recorded. The surface morphology of the nanoparticles was evaluated by scanning electron microscopy using JEOL JSM 7600F Scanning Electron Microscope. Analysis of drug loading, encapsulation efficiency and in-vitro drug release was done by a reversed phase high performance liquid chromatography (RP-HPLC) using the Agilent- Hewlett Packard 1100 Series High Performance Liquid Chromatography System equipped with Eclipse plus C18 (4.6 × 150 mm, 5 μm particle size) column kept at 25°C using 50:50 (acetonitrile:water) as mobile phase at a flow rate of 1mL/min. The amount of aldehyde released from acetal hydrolysis studies of blank nanoparticles was measured by U.V. spectrometer (UV-2401 PC, Shimadzu). The nanoparticles were lyophilized using Labconco Freeze dry system/Freezone 4.5. The drug release studies were carried out using a Labquake Shaker capable of 360° rotation and maintained at 37°C in endotherm laboratory oven (Fischer scientific, USA). Cell viability was determined using the CellTiter-Glo® luminescent cell viability assay (CellTiter®-Glo Luminescent Cell Viability Assay, Technical Bulletin, TB 288; Promega). Luminescence for determination of cell viability (%) was measured using the Labsystems Luminoskan RT microplate reader. In-vitro uptake of rhodamine-123 loaded nanoparticles was determined using the confocal laser scanning microscope (Nikon configuration D-Eclipse C1 with a TE2000 microscope, Nikon Instruments, Inc.).
Synthesis of di(2-methacryloyloxyethoxy)-[2,4- dimethoxyphenyl]methane (Bisacrylate Acetal Croslinker)
The reaction of 2,4-dimethoxybenzaldehyde (1.9982 g, 12.02 mmol), HEMA (6 ml, 49.47 mmol), p-toluenesulfonic acid monohydrate (0.3625 g, 1.906 mmol) and 4A° molecular sieves (5.5 g) was carried out in 25 ml of anhydrous dichloromethane (anhyd. DCM) at room temperature under nitrogen gas for 30 minutes and for another 24 hours without nitrogen gas. Then, the reaction mixture was quenched with triethylamine (2.1 ml, 15.1 mmol) at 0°C for 30 minutes, filtered, washed with dichloromethane and evaporated to give a crude liquid product. The crude product was purified by column chromatography using silica gel as a stationary phase and nhexanes/ ethyl acetate (50:50) with triethylamine (1% v/v) as the mobile phase.
Characterization of Acetal crosslinker
1H NMR and 13C NMR analyses of synthesized acetal crosslinker in CDCl3 were carried out on a Bruker AVANCE 400 MHz NMR spectrophotometer. Reported chemical shifts (ppm) are relative to residual CDCl3 peak and coupling constants were reported in Hz. 1H NMR spectra were recorded at 400 MHz while 13C NMR spectra were recorded at 100 MHz. FT-IR spectrophotometric analysis of crosslinker was carried out using a Perkin Elmer Spectrum 100FT-IR spectrometer enabled with attenuated total reflectance (ATR) technology. This equipment does not require the preparation and use of potassium bromide pellets. The molecular weight of the crosslinker was determined by high resolution mass spectroscopy. Analyte was dissolved in Acetonitrile (50%) +Trifluoric acid (0.1%) and injected into Thermo LTQ Orbitrap XL tandem Mass Spectrometer by syringe pump, using Nano Electro Spray Ionization (NSI). Mass spectrometer was operated in FT positive ion mode with resolution 30,000.
Hydrolysis studies of the crosslinker
Stock solutions (5 mg/ml) of the crosslinker were prepared in tetrahydrofuran (THF). 20 μL of the stock solution was added to 3 ml of PBS buffer (pH 7.4) or acetate buffer (pH 5.0). The hydrolysis of the crosslinker was investigated at pH 7.4 and pH 5.0 after incubation of the solutions at 37°C, using a UV-V is spectrophotometer (UV-2401PC). UV absorbance of the solutions was measured at pre-determined time points at a wavelength of 275 nm. This wavelength number is the λmax of 2,4- dimethoxybenzaldehyde which is the hydrolysis product of the crosslinker. Complete hydrolysis of the crosslinker was determined by taking the maximum absorbance after 24 hours of hydrolysis of the crosslinker at 37°C. The percent hydrolysis of the crosslinker at each pH value was expressed as a percentage of the maximum absorbance [15,17,18,25].
Preparation of blank and docetaxel-loaded crosslinked nanoparticles by dispersion polymerization
The blank and docetaxel-loaded nanoparticles were synthesized by dispersion polymerization technique [17,19-24], using n-butyl acrylate monomer, poly (ethylene glycol) n monomethyl ether mono methacrylate (MW of PEG Block=1,000) (a comonomer and a steric stabilizer), synthesized acetal crosslinker and benzoyl peroxide/Nphenyldiethanolamine (BPO/N-PDEA) as a co-initiator system.
For the synthesis of blank nanoparticles, 1.146mL, 7.993 mmol of n-butyl acrylate monomer, 0.1002 g, 0.1002 mmol of PEG-MMA and 0.309 g, 0.756 mmol of the crosslinker were dissolved in methanol: water solvent system (10:3) in a three necked round bottom flask. 0.0405 g, 0.1672 mmol of BPO and 0.0303 g, 0.1672 mmol of N-PDEA were injected at predetermined intervals through the rubber closure under continuous flushing with nitrogen gas and stirring for 18 hours. The resulting suspension was further purified by introducing it into a dialysis tube with a 12,000-14,000 Dalton molecular weight cut-off (Spectra/Por®CE), and dialyzed against phosphate buffer. The dialysis medium was changed at selected time intervals with fresh phosphate buffer. After dialysis, the nanoparticles were freeze dried using Labconco Freeze dry system/ Freezone 4.5 (Labconco, Kansas City, MI) for 24 hours and then stored in the refrigerator at 4°C. The method of preparation for drug-loaded nanoparticles was the same as for blank nanoparticles except that 10mg of docetaxel was added during the nanoparticle synthesis and also the drugloaded nanoparticles were purified by centrifugation using an ultracentrifuge. The centrifugation was carried out at 4°C and 19,000 RPM for 10 minutes. After centrifugation, the supernatant was discarded and the nanoparticle pellet was dispersed in deionized distilled water and lyophilized for 24 hours and stored at 4°C.
Characterization of blank and docetaxel-loaded nanoparticles by Scanning Electron Microscopy
Different dilutions of nanoparticle suspension in distilled water were placed on a carbon tape affixed to a specimen (aluminum) stub (SPI Supplies, Inc.) and dried in a vacuum oven for 24 hours. The samples were gold coated with Hummer sputtering machine for 2 minutes under argon atmosphere to improve conductivity The samples were then observed using JEOL JSM 7600F scanning electron microscope. A 5 kV accelerating voltage and secondary electron mode was used with a working distance of 12 mm for the morphological characterization. The images were taken at different magnifications.
Particle size and size distribution analysis
Particle size distribution and average particle size of nanoparticles were determined by dynamic light scattering (DLS) technique. A known weight of freeze dried particles was dispersed in 5mL of filtered distilled deionized water; then the suspension was placed in a cuvette (Malvern, Inc.). Particle size was determined at 25°C using a Malvern Zetasizer Nano ZS (Malvern Instruments, Inc., USA). The mean of three measurements was recorded. The particle size distribution is expressed as polydispersity index (PDI). The lower the value, the narrower the size distribution or the more uniform is the nanoparticle sample. A PDI of 0 indicates an ideal monodisperse formulation; while a PDI of 1 indicates large variations in particle size.
Zeta potential determination
A known weight of freeze-dried nanoparticles was dispersed in filtered distilled deionized water as described for particle size analysis. Following filtration through an Acrodisc® syringe filter with a 5 μm Versapor® membrane (Pall Corporation), 1.5 mL of the resulting suspension was diluted with 1.5 mL of filtered distilled water and mixed by vortexing. Then this suspension was placed in a capillary zeta potential cell (Malvern, Inc.) and analyzed using Malvern Zetasizer Nano ZS (Malvern Instruments, Inc., USA). The mean of three measurements was recorded.
Drug loading and encapsulation efficiency
Preparation of calibration curves for docetaxel: Analyses of drug loading, encapsulation efficiency and in-vitro drug release were done by reversed phase high performance liquid chromatography (RP-HPLC) using the Agilent-Hewlett Packard 1100 Series High Performance Liquid Chromatography System equipped with Eclipse plus C18 (4.6 × 150 mm, 5 m particle size) column kept at 25°C using 50:50 (acetonitrile:water) as the mobile phase at a flow rate of 1mL/min. Calibration curves were constructed for docetaxel by linear regression analysis of the peak area versus drug concentration. The stock solution was prepared by dissolving 5 mg of docetaxel in 10mL of acetonitrile to give a final concentration of 0.5 mg/mL. From the stock solution, dilutions were made to prepare solutions with concentration ranging from 2.5, 5, 10, 15, 20 to 25 μg/mL. The diluent used for the preparation of the solutions was acetonitrile: water (50:50). Linearity was demonstrated (R2=0.9994). Ultravioletvisible detection was performed using an HP photodiode array detector. The eluents were monitored at 230 nm. No interfering substances eluting at the retention time for docetaxel were observed for the blank nanoparticles.
Drug loading (DL) was determined by dissolving a known weight of docetaxel-loaded nanoparticles (ANp) in 2 mL of dichloromethane. The solution was filtered through a 0.45 m syringe filter and the amount of docetaxel dissolved in the solution (ASol) was quantified by reversed phase high performance liquid chromatography (RP-HPLC) method. Briefly, 20 μL of the filtered solution was injected into HPLC kept at 25°C using 50:50 (acetonitrile: water) as the mobile phase at a flow rate of 1 mL/min. The percent DL was calculated from the equation (1) as shown below [19,24]:
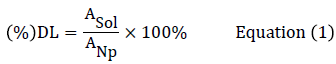
Encapsulation efficiency (EE) was determined by quantifying the amount of docetaxel in the supernatant (ASup) after ultracentrifugation by RPHPLC method with the assumption that the rest of the drug used for nanoparticle preparation had been encapsulated into the nanoparticles.The supernatant solution was filtered through a 0.45 μm syringe filter and 20 μL of the filtered solution was injected into HPLC instrument for the analysis of the amount of docetaxel in total volume of supernatant. The initial amount of drug used in the synthesis of nanoparticles was designated as (APrep). The amount in the supernatant was designated as (ASup). The Encapsulation efficiency (EE) was determined from the equation (2) as below:
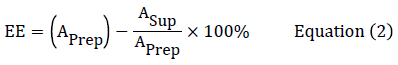
Hydrolysis studies of blank nanoparticles
The particle cleavage kinetics of blank nanoparticles synthesized with acetal crosslinker was determined using a UV-V is spectrophotometer (UV-2401 PC, Shimadzu Corporation), by measuring the concentration of the cleavage by-products, (2,4 dimethoxybenzaldehye) formed upon hydrolysis of the acetal crosslinker from the nanoparticles at 275 nm. The dialysis bag containing a known weight of the blank nanoparticles in 2 ml of buffer was immersed in a 15 ml centrifuge tube containing 12 mL of either pH 5 or pH 7.4 buffers, and the tubes were clamped to a Labquake® shaker capable of 360° rotation maintained at 37°C in an endotherm laboratory oven (Fischer Scientific, USA). At predetermined time intervals, 1 mL of the dialysis medium was removed for the measurement. The volume of the dialysis medium was retained by adding fresh 1 mL of the buffer solution at relevant pH after each sampling to maintain constant volume. The experiments were performed in triplicate [17,26,27].
In-vitro drug release studies
In-vitro drug release was measured by a dialysis method [19-24]. A known amount of docetaxel-loaded nanoparticles was suspended separately in 2 mL of phosphate buffered saline (PBS-pH 7.4) and acetate buffer (pH 5.0) and placed in separate dialysis bags with a 12,000-14,000 Da molecular weight cut-off (Spectra/Por®CE). The dialysis bags were then placed in 15 mL centrifuge tubes containing a known volume of PBS 7.4 and pH 5 acetate buffers. The tubes were clamped to a Labquake® shaker capable of 360° rotation and maintained at 37°C in an endotherm laboratory oven (Fischer Scientific, USA). At specific time intervals, an aliquot of the release medium was taken and replaced with the same amount of fresh medium each time to maintain constant volume and sink conditions. The amount of docetaxel released in each medium was quantified by RP-HPLC analysis method. The release profile was determined by plotting a graph of (%) cumulative amount of docetaxel released versus release time. The concentration of released docetaxel was corrected for the dilution due to the addition of fresh medium.
Biological studies on cancer cells
Cytotoxicity Studies on PC3 and LNCaP prostate cancer cell lines
The docetaxel-loaded nanoparticles were used for in-vitro cytotoxicity studies. Blank nanoparticles were used as a comparison. Prostate cancer cells (PC3 and LNCaP) were seeded in 96-well plates (New Corning Costar 3903, opaque, white, flat bottom) at a seeding density of 5000 and 10,000 cells/well /0.1 ml respectively. The medium used was RPMI 1640 containing 10% FBS and the cells were placed in an incubator and allowed to attach for 48 hours. After 48 hours of incubation at 37°C, the old media was aspirated off and replaced with fresh medium containing different concentrations of docetaxel solution or docetaxel-loaded nanoparticles at an equivalent concentration of drug ranging from 0.5 to 100 nM. Following drug treatment, at different time intervals (24-, 48- or 72 hours), assay plates were removed from the incubator, allowed to equilibrate to room temperature for about 30 minutes, and 100 μL of CellTiter-Glo® assay reagent, equilibrated to room temperature, was added to each well. Plates were shaken for 2 min to mix the contents of the wells using an orbital plate shaker. Following 10 minute incubation at ambient temperature, luminescence was determined using a Labsystems Luminoskan RT microplate reader. Control cells were treated with medium only, medium with dimethyl sulfoxide (DMSO) and medium containing blank nanoparticles at the highest concentration of docetaxelloaded nanoparticles tested. For data analysis, the measured luminescence from cells treated with docetaxel-loaded nanoparticles and docetaxel solution was normalized to the luminescence from cells with no treatment (medium only) which was set at 100% and used as a control. Results are presented as percent cell viability and as the mean ± S.D. of 4 replicates per concentration tested.
Cell internalization studies using confocal laser scanning microscope
In order to study cellular uptake of nanoparticles in-vitro or in-vivo, the use of fluorescently or radioactively labeled nanoparticles is the most common experimental approach. Fluorescent labeling was chosen in the present research work. Fluorescent labeling makes cellular uptake of nanoparticles readily detectable by fluorescence microscopy or confocal laser scanning microscopy (CLSM). Rhodamine-123 is a lipophilic fluorescence probe that has been widely used to study cellular uptake of nanoparticles. In this study rhodamine-123 loaded nanoparticles were synthesized and used for cell uptake studies using a confocal laser scanning microscope [23,28,29]. Rhodamine-123 labeled nanoparticles were prepared in the same way as the docetaxel-loaded nanoparticles. Instead of docetaxel, rhodamine-123 (Invitrogen, USA) was incorporated into the nanoparticles for fluorescence detection. PC3 cells were seeded (30,000 cells/1 mL/dish) in glass bottom microwell dishes (35 mm- dish diameter, MatTek Corporation, USA) and incubated for 48 hours. After 48 hours, old medium was removed and cell monolayers were then treated with 1 mL of rhodamine-123 loaded nanoparticles (250 μg/mL) dispersed in the serum free medium for 2 hours and 4 hours. After specified time intervals, the cells were stained with Hoechst® 33342 dye (Invitrogen, USA) for 1 hour and then the medium was aspirated off. The dishes were then rinsed three times with phosphate buffered saline (PBS buffer) to remove all the free nanoparticles that were not internalized into the cells, and stained with CellMask™ Orange Plasma membrane stain [23]. The cells were then viewed and imaged by a confocal laser scanning microscope (Nikon configuration D-Eclipse C1 with a TE2000 microscope, Nikon Instruments, Inc.). The laser excitation and emission bandpass wavelengths were 408 nm and 450 ± 35 nm for blue filter, 488 nm and 515 ± 30 nm for green filter and 543nm and 605 ± 75 nm for red filter. The nucleus stained with Hoechst® 33342 dye showed blue color, rhodamine-123 labelled nanoparticles appeared green and the cell membrane stained with CellMask™ Orange Plasma membrane stain showed red color.
Results and Discussion
Synthesis of Benzaldehyde Bisacrylate Acetal crosslinker:
The main aim in the synthesis of acid- labile crosslinker is to achieve the desired pH-responsive behavior, i.e. rapid cleavage in slightly acidic pH such as those present in intracellular vesicles of the cells (pH4.5-6.8), and stability for long time in the blood, i.e. pH 7.4. The acetals synthesized from p-substituted benzaldehydes have been shown by the previous studies [15,30] to be particularly well suited to this purpose as their acidlabile behavior can be tailored by introducing different substituents at the para-position of the benzaldehyde. Thus, the use of the crosslinkers containing p-substituted benzaldehyde acetals as an intrinsic acidresponsive element provides an easy and versatile way to incorporate and engineer the acid-cleavability to the polymeric colloidal systems for the site-specific drug release [30].
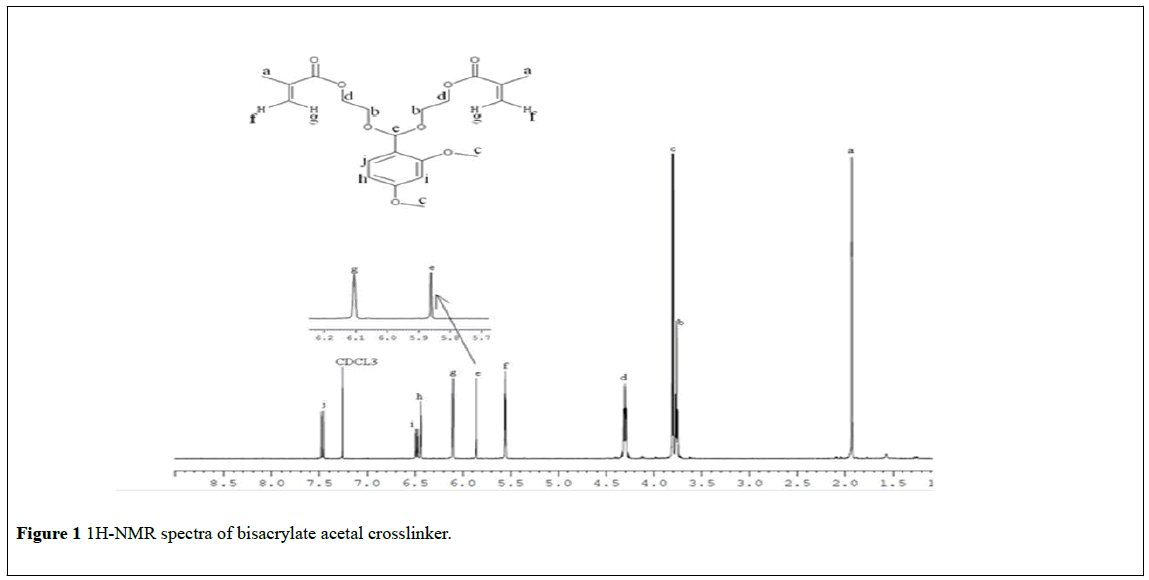
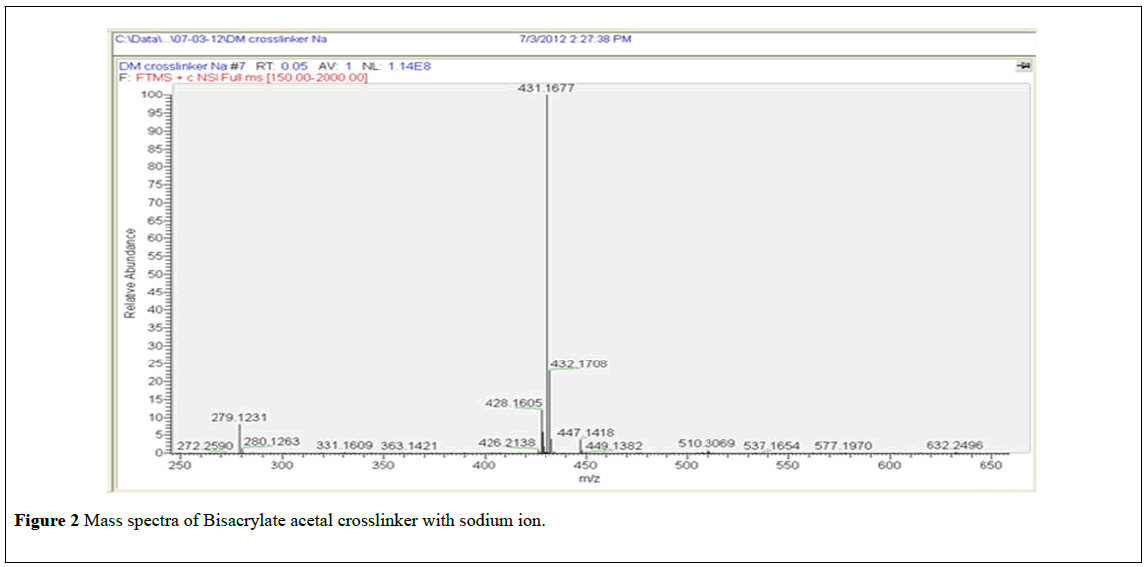
The benzaldehyde (2,4-dimethoxy benzaldehyde) was reacted with 2- hydroxyethyl methacrylate (HEMA) in the presence of p-toluenesulfonic acid to form bisacrylate acetal crosslinker. The chemical structure of the crosslinker is as shown below in Scheme 1. The formation of acetal crosslinker was confirmed from the 1H NMR, 13C NMR, FT-IR spectra and high resolution mass spectroscopy. 1H-NMR spectra showed the presence of singlet acetal protons (δ 5.85 ppm). Also, there was no aldehyde peak observed in the 1H-NMR spectra of the crosslinker, which further confirmed the formation of acetal. All the other peaks and chemicals shifts were as expected (Figure 1). The IR spectrum of crosslinker showed presence of alkane C-H stretch at 2956 cm-1. The sharp absorption at 1715 cm-1 (C=O stretch) and multiple sharp bands of C-O stretch from 1000-1300 cm-1 confirm the presence of an ester group. The presence of a C=C stretch at 1637 cm-1 indicates the C=C stretch from the vinyl groups of the crosslinker. Medium to weak multiple bands between 400-1600 indicates C=C stretch from an aromatic ring. (Figure not shown). The result of high resolution mass spectroscopy was in agreement with the expected mass of the crosslinker (Figure 2). The 13C NMR spectrum of the crosslinker confirms the structure (Figure not shown). The crosslinker was a colorless viscous liquid with yield of 36%. 1H NMR (400 MHz, CDCl3 ppm): 1.93 (t, 6H, J=1.3 Hz), 3.75 -3.78 (m, 4H), 3.79 (s, 3H), 3.81 (s, 3H), 4.26 - 4.35 (m, 4H), 5.55 - 5.56 (m, 2 H), 5.86 (s, 1H), 6.10 (t, 2H, J=1.3 Hz), 6.44 (d, 1H, J=2.36 Hz), 6.476.49 (dd, 1H, J=8.4 Hz, 2.4 Hz), 7.47, (d,1H, J=8.4 Hz). 13C NMR (100 MHz, CDCl3 ppm): 18.095, 55.241, 55.402, 63.409, 63.708, 97.266, 98.411, 104.019, 125.362, 128.170, 136.156, 158.114, 161.196, 167.155. HR-MS (theoretical mass for C21H28O8: 408.44 g/mol, measured mass: 408.17 g/ mol).
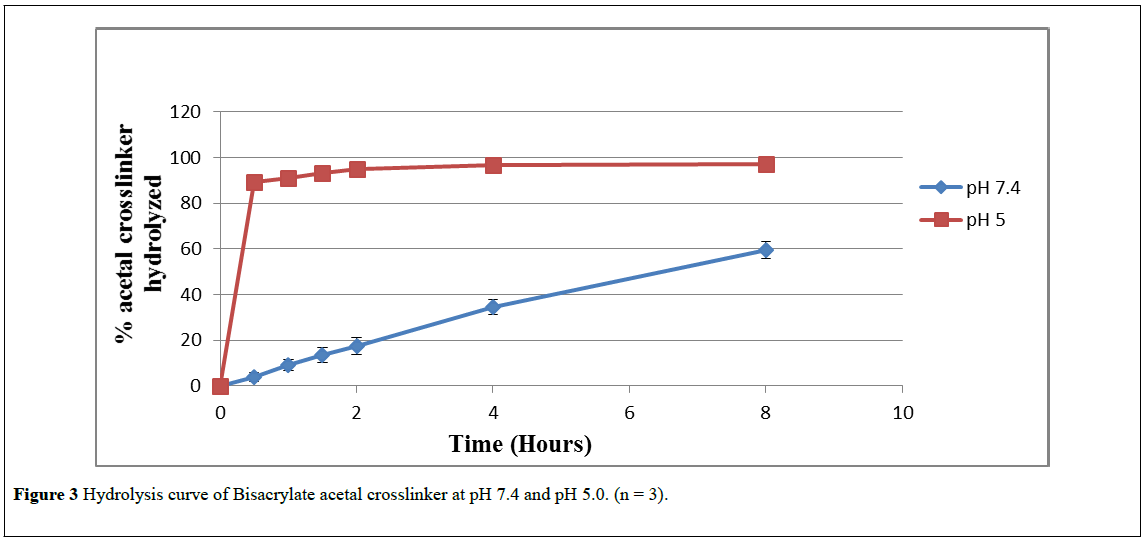
Hydrolysis of Bisacrylate Acetal crosslinkers
Figure 3 shows the plot of percentage of acetal crosslinker hydrolyzed versus time in pH 7.4-buffer and pH 5.0 buffer. The rate of hydrolysis of the crosslinker is faster at pH 5.0 as compared to pH 7.4. Akala and his coworkers have explained in detail the mechanism of acid-catalyzed hydrolysis of para-methoxybenzaldehyde acetal to paramethoxy benzaldehyde and 2-hydroxyethylacrylate in the preliminary work that preceeded this manuscript [17]. The acceleration of the hydrolysis kinetics of the crosslinker from from pH 7.4 to pH 5.0 is expected because the rate of hydrolysis of benzaldehyde acetals is proportional to the hydronium ion concentration which increases 250 fold between pH 7.4 and pH 5.
Characterization of nanoparticles
Scanning electron microscopy studies
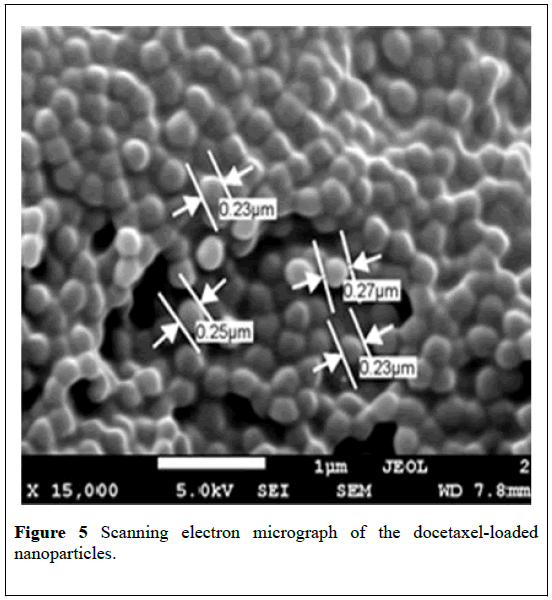
The formation of nanoparticles was confirmed by scanning electron microscopy (SEM) as shown in Figures 4 and 5. The images show that spherical nanoparticles were obtained. The images demonstrated a narrow particle size distribution, in support to the findings from particle size analysis as shown in Table 1.
| Nanoparticle | Particle Size (nm) | PDI | Zeta Potential (mV) | Encapsulation efficiency (%) | Loading Efficiency (%) |
|---|---|---|---|---|---|
| Blank nanoparticles | 205.87 ± 13.15 | 0.37 ± 0.05 | -21.77 ± 1.33 | - | - |
| Docetaxel-loaded nanoparticles | 265 ± 1.96 | 0.23 ± 0.003 | -30.13 ± 0.51 | 90.48 ± 0.31 | 1.79 ± 0.24 |
Table 1: Average particle size, PDI, zeta potential, encapsulation efficiency and loading efficiency of nanoparticles (n = 3).
It was observed that the size of the nanoparticles increased slightly after incorporation of docetaxel into the nanoparticles. SEM images showed the particle size is slightly lower than that obtained from dynamic light scattering (DLS) experiment.
The tendency to shrink and collapse while the nanoparticles are in dry state may contribute to this difference with the DLS results [31,32]. The particle size from the DLS measurement was the size of the particles plus an aqueous layer that surrounds the particles and moves together with the particles [33]. Moreover, the possibility of slight swelling of the nanoparticles while in an aqueous environment for DLS measurement cannot be ruled out. Hence, the particle size observed from DLS experiments was slightly larger than that obtained from SEM.
Particle size
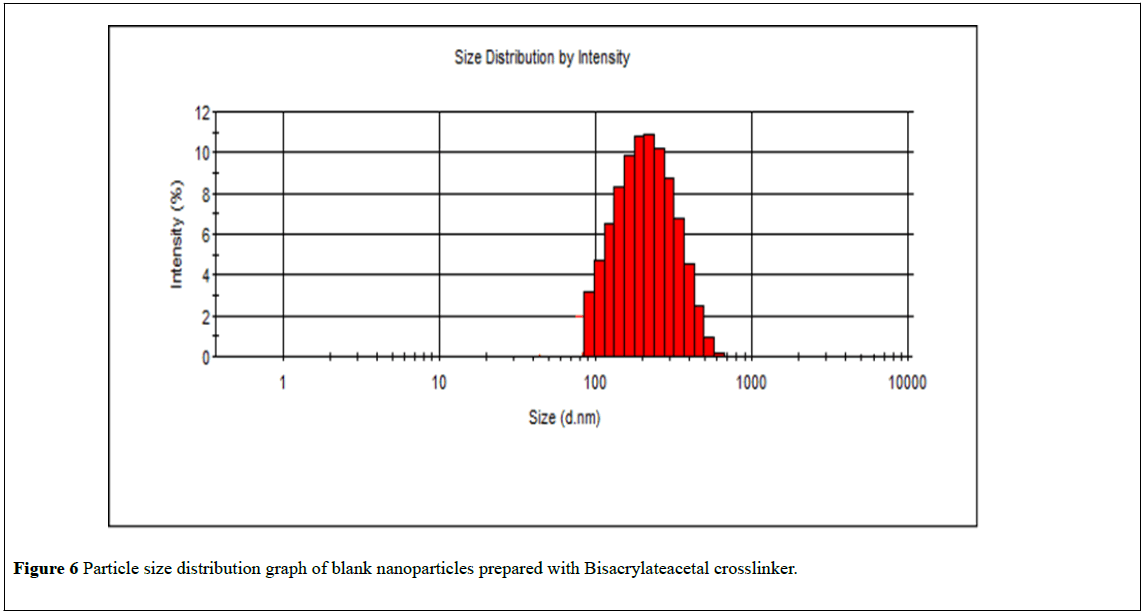
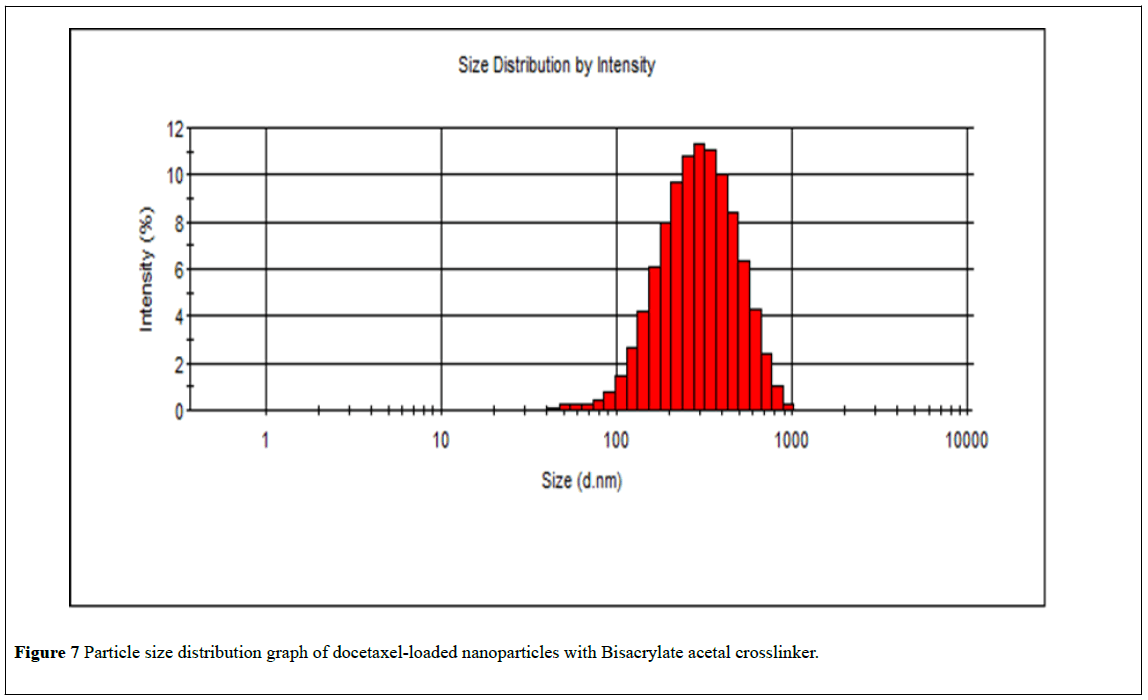
Physicochemical characteristics of the nanoparticles such as particle size and surface charge, play an important role in determining their in-vitro drug release, as well as their in-vivo pharmacokinetics and biodistribution, and hence the therapeutic efficacy of the encapsulated drug [34,35]. When large colloidal particles are administered intravenously they are rapidly taken up by RES while small particles and those with a hydrophilic surface show slow clearance rate [36]. In this study, the hydrophilic surface was provided by PEG-MMA, which has been shown to give longcirculating nanoparticles. The particle size distribution curves of nanoparticles are shown in Figures 6 and 7. Thus it appears that the encapsulated docetaxel contributed to the enlargement of the particles size. The increase in size can possibly be attributed to new interactions between docetaxel and n-butyl acrylate monomer and crosslinkers. The addition of the drug in the formulations produces an expansion of the polymeric matrix, thereby increasing particle size. Docetaxel-loaded nanoparticles had particle size 265 ± 1.96 nm, which is in size range favoring cellular uptake of nanoparticles [34].
Figure 6: Particle size distribution graph of blank nanoparticles prepared with Bisacrylateacetal crosslinker.
Figure 7: Particle size distribution graph of docetaxel-loaded nanoparticles with Bisacrylate acetal crosslinker.
Zeta potential
Zeta potential is the difference in the electrical charge developed between the dense layers of ions surrounding the particles and the charge of the bulk of the suspended fluid surrounding the particle. Zeta potential gives information about the overall surface charge of the particles. Thus, the measurements of zeta potential may indicate colloidal stability of nanoparticles, as interactions between particles play an important role in determining colloidal stability. The use of zeta potential measurements to predict stability attempts to quantify such interactions. Most nanoparticles have a tendency to aggregate which may lead to precipitation that could prove dangerous if those particles are injected intravenously. Since most aqueous colloidal systems are stabilized by electrostatic repulsion, the larger the repulsive forces between particles, the less likely they will come closer together and form an aggregate. Therefore, it is important to know the surface charge, which directly controls aggregation behavior of the particles in the blood [37]. It is also an important factor in envisaging the interaction of nanoparticles in-vivo with the biological cell membrane. In addition, from the zeta potential measurement, we can roughly know the dominant component on the particle’s surface [38]. It has been reported that a higher absolute value of the zeta potential indicates a more stable suspension, and a lower value indicates colloid instability, which could lead to aggregation of nanoparticles. In general, higher the absolute value of zeta-potential (>15 mv) is, the stronger the electrostatic repulsion between particles. [39]. Zeta potential values obtained in this study range from 21.77 ± 1.33 to -30.13 ± 0.51 (Table 1). Puri, et al. [17] has reported that nanoparticles with the smallest absolute value of zeta potential have the smallest particle size. It has also been reported that PEG outer shell is capable of lowering the negative surface charge to a certain degree, resulting in reduction of cell repulsion to the nanoparticles [40,41]. It can be inferred that the smallest nanoparticles obtained in this work have the maximum PEG coating on the outer surface, and hence the smaller negative zeta potential value.
Drug loading and encapsulation efficiency
Drug loading of nanoparticles is an important factor in the nanoparticle drug formulation since high drug loading implies that less amount of the nanoparticle is needed for a given dose of treatment [42]. The encapsulation efficiency and loading efficiency of the nanoparticles are as shown in Table 1. The encapsulation efficiency of nanoparticles is 90.48 ± 0.31%; while the loading efficiency of nanoparticles is 1.79 ± 0.24%.
Hydrolysis of blank nanoparticles
The pH-responsiveness of nanoparticles was characterized chemically by monitoring the hydrolysis of the acetal crosslinker in the nanoparticles. When exposed to aqueous acidic medium, the nanoparticles underwent acid-catalyzed hydrolysis and released free acetal, which further dissociated to an aldehyde and alcohol [17]. The aldehyde released has an ultraviolet (UV) absorbance peak at a specific wavelength when in solution but not in the acetal form when still attached to the polymer. This easily allows monitoring of the acetal hydrolysis rates using ultraviolet/ visible (UV/Vis) spectroscopy [15,27].
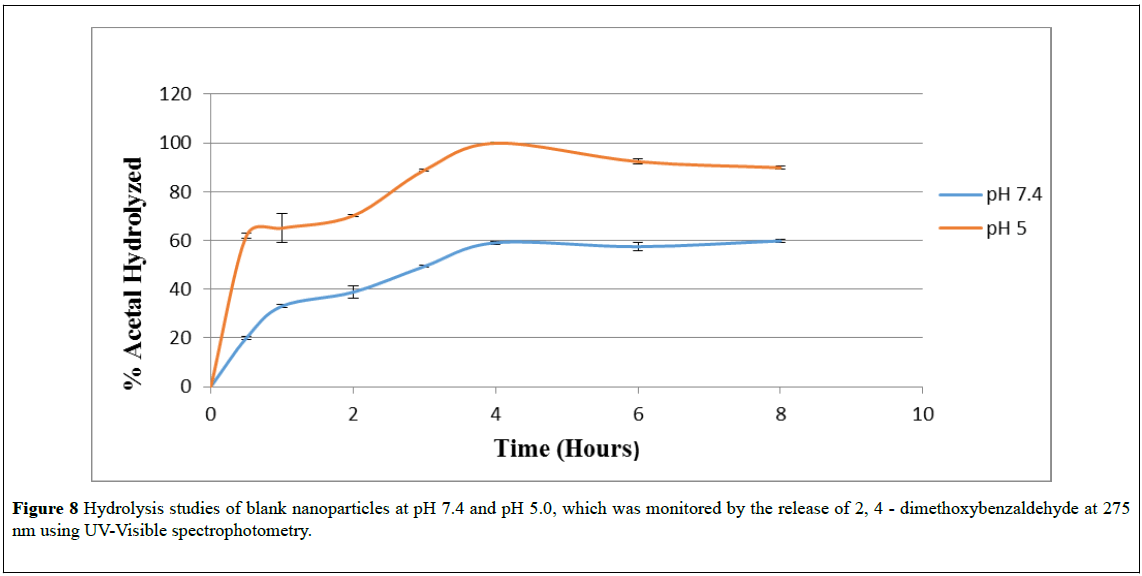
Nanoparticle degradation was studied at physiological pH (7.4) and lysosomal pH (5.0). Specifically, the amount of the aldehyde, the byproduct of acetal degradation (2,4-dimethoxy benzaldehyde monitored at 275 nm) released from nanoparticles at different time intervals was determined. The hydrolysis curves of the nanoparticles are shown in Figure 8. The results show that the hydrolysis rate of the acetals from the nanoparticles is pH dependent. The rate and the extent of hydrolysis of the acetal from the nanoparticles were faster and higher respectively at pH 5 as compared to pH 7.4.
Figure 8: Hydrolysis studies of blank nanoparticles at pH 7.4 and pH 5.0, which was monitored by the release of 2, 4 - dimethoxybenzaldehyde at 275 nm using UV-Visible spectrophotometry.
Drug release studies
The key feature of the pH-responsive nanoparticles for cancer therapy is their ability to respond rapidly to the acidic environment in the endosomal and lysosomal compartments by releasing the encapsulated bioactive agent(s). The pH-dependent cleavage behavior of the particles is due to the hydrolysis of the acetal bonds crosslinking poly (n-BA) chains in the particle bulk structure. Upon cleavage of the acetal, the polymer nanoparticle changes from a hydrophobic structure to an open hydrophilic one. Thus water enters the structure and causes the nanoparticles to degrade and release its contents [43].
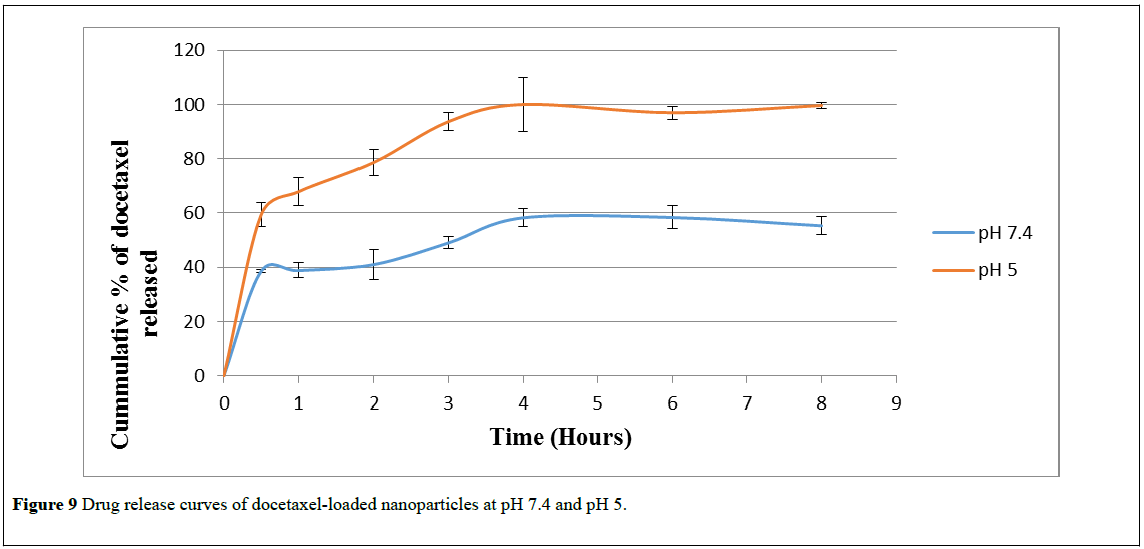
The release kinetics of docetaxel from the particles (Figure 9) is very similar to the hydrolysis kinetic of blank nanoparticles (Figure 8). At 3 hours, the cumulative amount of docetaxel released from nanoparticles was 94.72% at pH 5.0; while the nanoparticles were fairly stable at pH 7.4 over a period of 8 hours when compared to pH 5.0. This similarity in the results of the hydrolysis of blank nanoparticles and drug release studies support the position that the release of docetaxel occurs due to the pHdependent cleavage of acetal bonds in the polymeric matrix. The mechanism of hydrolysis kinetics of the acetal bonds and the particle cleavage may be explained briefly as follows. In an acidic solution, the acid protonates one oxygen of the acetal, creating a weak base of the relevant neighboring group of the carbonyl carbon. This makes it easier for water to attack the carbonyl carbon resulting in the cleavage of the acetal bonds to the relevant aldehyde and alcohol. Since there are almost 250 fold increase in the concentration of hydronium ions in the solution at pH 5 compared to the solution at pH 7.4, the protonation of the carbonyl oxygen and thus the hydrolysis of the acetal is expected to accelerate proportionally when the pH is decreased from pH 7.4 to pH 5. As a result of the hydrolysis of the acetal bonds that crosslink the particle network, disintegration of the particles occurs and the drug is released [17].
Figure 9: Drug release curves of docetaxel-loaded nanoparticles at pH 7.4 and pH 5.
Cytotoxicity studies
The in-vitro cytotoxicity of docetaxel both as a free drug and when loaded into nanoparticles , at the same drug equivalent concentration of 0.5 to 100 nM, was evaluated by the CellTiter®-Glo Luminescent Cell Viability Assay using PC3 and LNCaP prostate cancer cell lines. LNCaP and PC-3 cells were chosen for this investigation to represent an androgen-sensitive and androgen insensitive prostate cancer cell line respectively [44]. Untreated PC3 and LNCaP cells were used as controls. Figures 10-12 and Figures 13-15, show the (%) viability of PC3 and LNCaP cells with free docetaxel solution and docetaxel-loaded nanoparticles after incubation for 24-, 48- and 72 hours respectively. Since no significant cell death was detected for blank nanoparticles for the duration of the assay tested as compared to medium-only treated controls, it appears that blank nanoparticles are nontoxic to PC3 and LNCaP cells, even at the highest nanoparticle concentration tested. This result shows that poly (n- butyl acrylate) nanoparticles have no intrinsic cytotoxicity towards PC3 and LNCaP cells. Hence, it can be assumed that the cell death caused by docetaxel-loaded nanoparticles was due to the docetaxel in the nanoparticles, rather than the nanoparticles themselves.
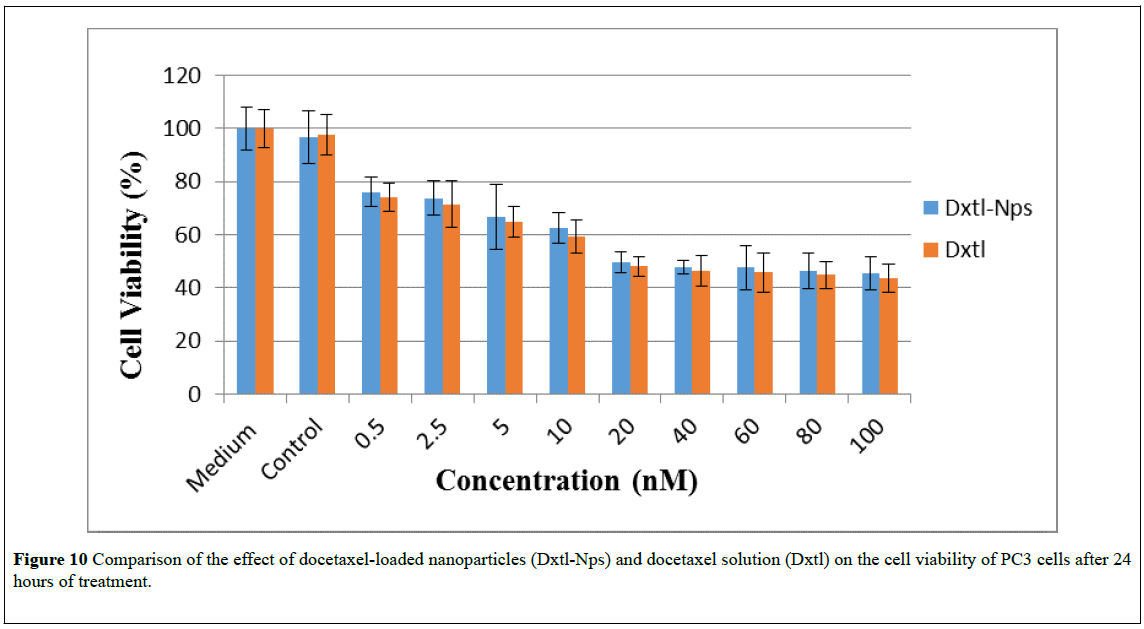
Figure 10: Comparison of the effect of docetaxel-loaded nanoparticles (Dxtl-Nps) and docetaxel solution (Dxtl) on the cell viability of PC3 cells after 24 hours of treatment.
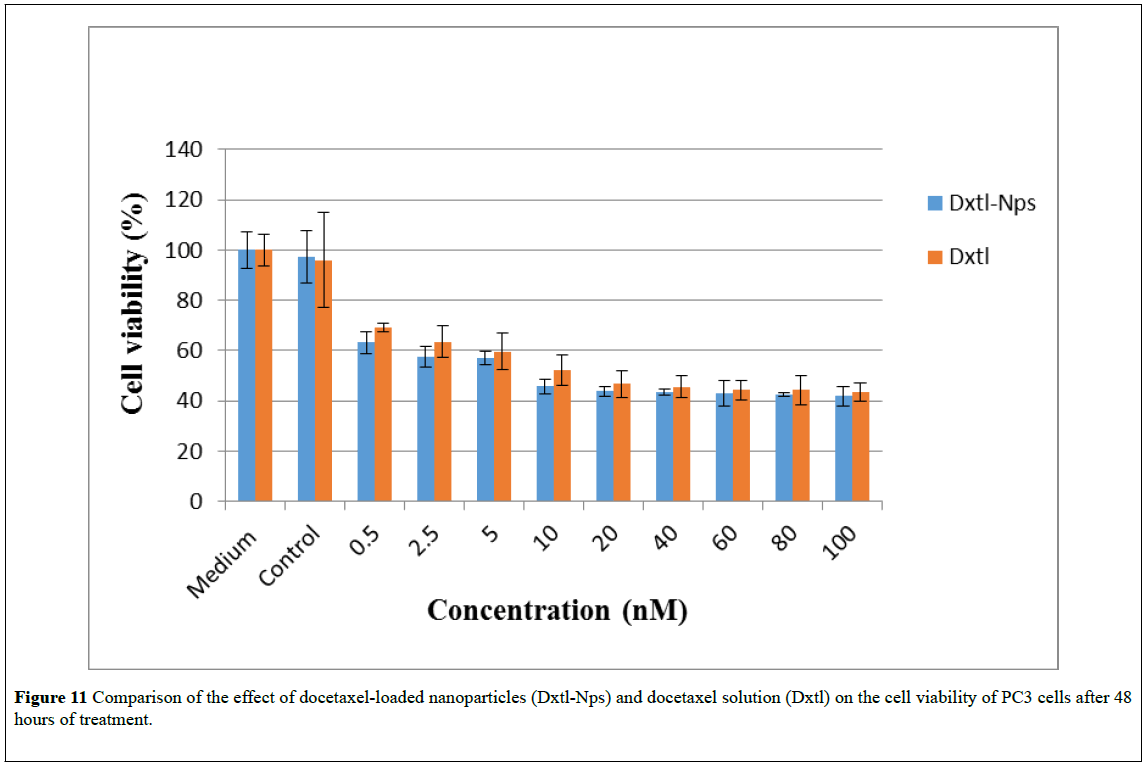
Figure 11: Comparison of the effect of docetaxel-loaded nanoparticles (Dxtl-Nps) and docetaxel solution (Dxtl) on the cell viability of PC3 cells after 48 hours of treatment.
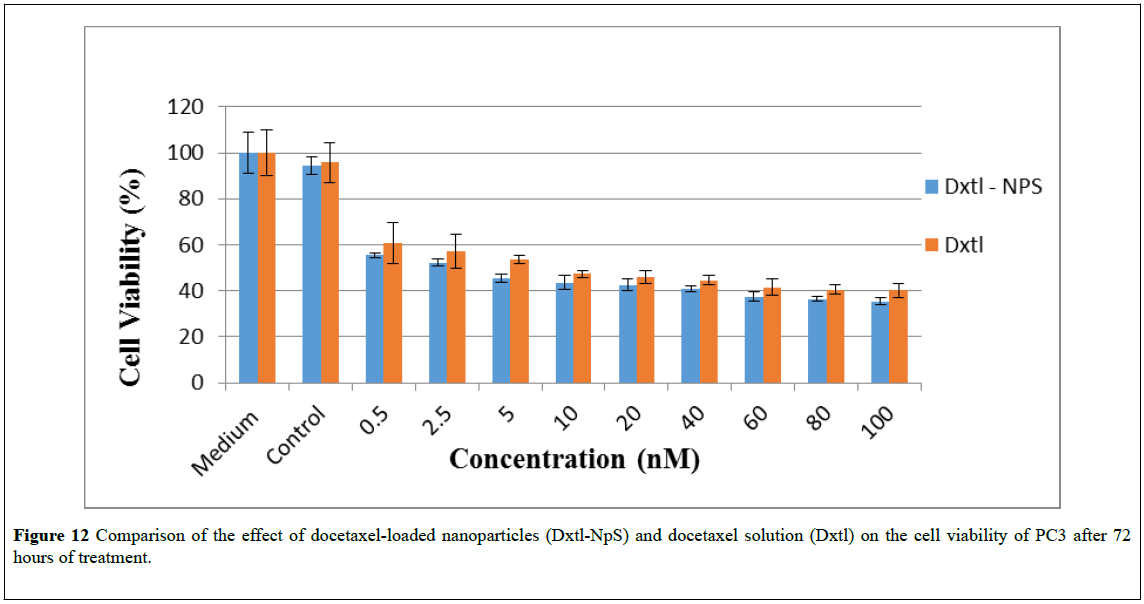
Figure 12: Comparison of the effect of docetaxel-loaded nanoparticles (Dxtl-NpS) and docetaxel solution (Dxtl) on the cell viability of PC3 after 72 hours of treatment.
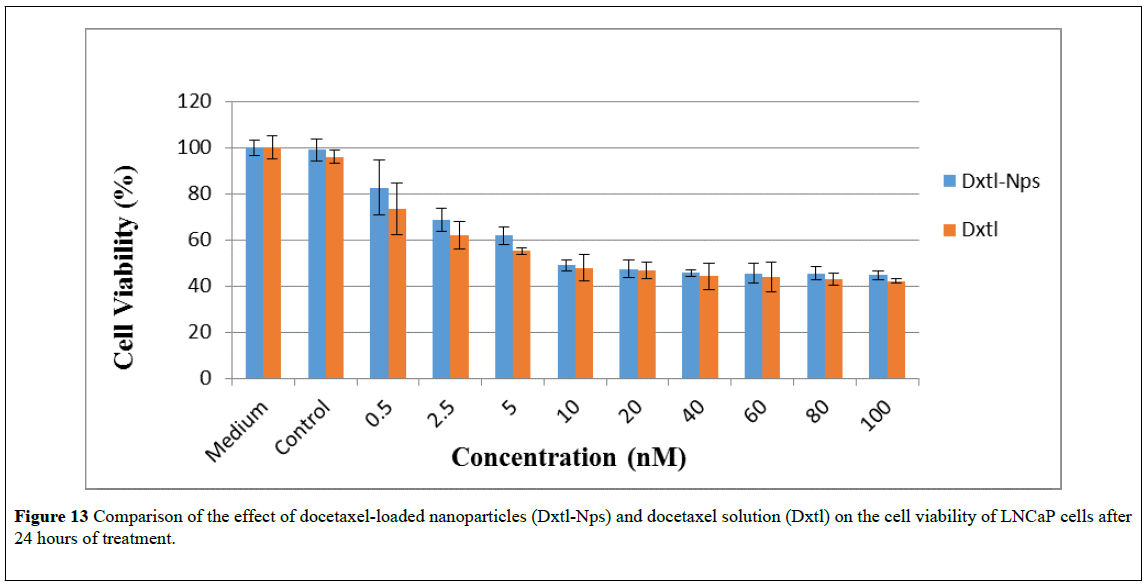
Figure 13: Comparison of the effect of docetaxel-loaded nanoparticles (Dxtl-Nps) and docetaxel solution (Dxtl) on the cell viability of LNCaP cells after 24 hours of treatment.
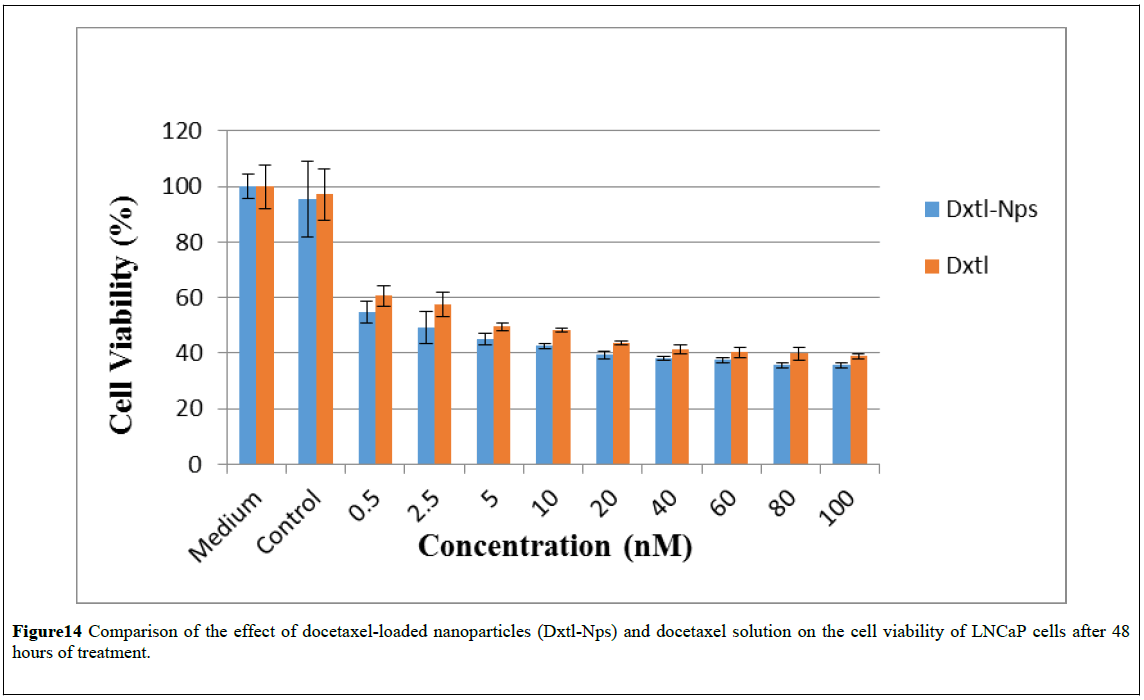
Figure 14: Comparison of the effect of docetaxel-loaded nanoparticles (Dxtl-Nps) and docetaxel solution on the cell viability of LNCaP cells after 48 hours of treatment.
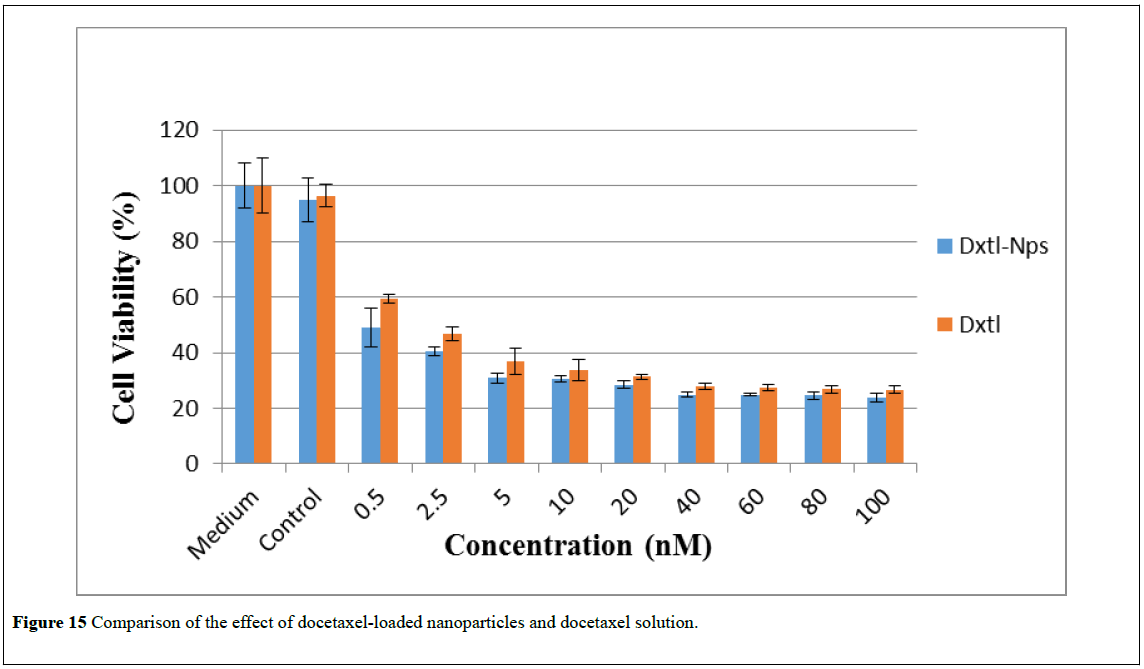
Figure 15: Comparison of the effect of docetaxel-loaded nanoparticles and docetaxel solution.
As shown in Figures 10 and 13, compared to free docetaxel, the docetaxel-loaded nanoparticles were slightly less effective against PC3 and LNCaP cells over the first 24 hrs. This observation may be due to the time required for the release of docetaxel from the nanoparticles. Drug release studies showed that docetaxel loaded into the nanoparticles was released completely in less than 24 hours. Thus it is only after 24 hours that the total amount of docetaxel loaded into the nanoparticles would be available in the medium. However, after 48 and 72 hours of exposure, (Figures 11, 12, 14 and 15), the nanoparticles containing docetaxel were as effective as or more effective than the free docetaxel PC3 (24 hours: docetaxel 42% and docetaxel-nanoparticles 48%); (48 hours: docetaxel 42% and docetaxel-nanoparticles 41%); (72 hours: docetaxel 42% and docetaxel-nanoparticles 36%) and LNCaP (24 hours: docetaxel 40% and docetaxel-nanoparticles 43%); (48 hours: docetaxel 38% and docetaxelnanoparticles 36%); (72 hours: docetaxel 25% and docetaxelnanoparticles 22%). The finding could be explained based on the mechanism of action of docetaxel, which requires cell division to operate. For longer incubation periods, a larger number of cells would enter G2 and M cell cycle phases, where docetaxel selectively works [4,45]. Hence, increasing the incubation time led to more cell death in PC3 and LNCap cells. Moreover, it was observed that an increase in the dose also resulted in an increase in cell death in both the cell lines. However, a plateau effect was observed above 40nM, above which there was no significant increase in cell death at all the time intervals tested. A similar result was obtained for paclitaxel-loaded PLA nanoparticles: no decrease in cell viability was found at higher paclitaxel concentrations (above 40 nM) [23]. The observation was explained as follows: the upregulation of the p53 protein by paclitaxel stimulates another gene to produce a protein (p21) that interacts with a cell division-stimulating protein (cdk2) [46]. When p21 is complexed with cdk2, cells cannot go to the next phase of cell division causing G1/G2 arrest and not mitotic arrest, thereby protecting against paclitaxel cytotoxicity because of the prevention of mitotic arrest. This reasoning agrees with the observation that if there is a G1 block, then cells would be unable to enter the M phase where paclitaxel exerts its effect [47]. A similar phenomenon may be operating in the cytotoxicity of docetaxel at high concentrations.
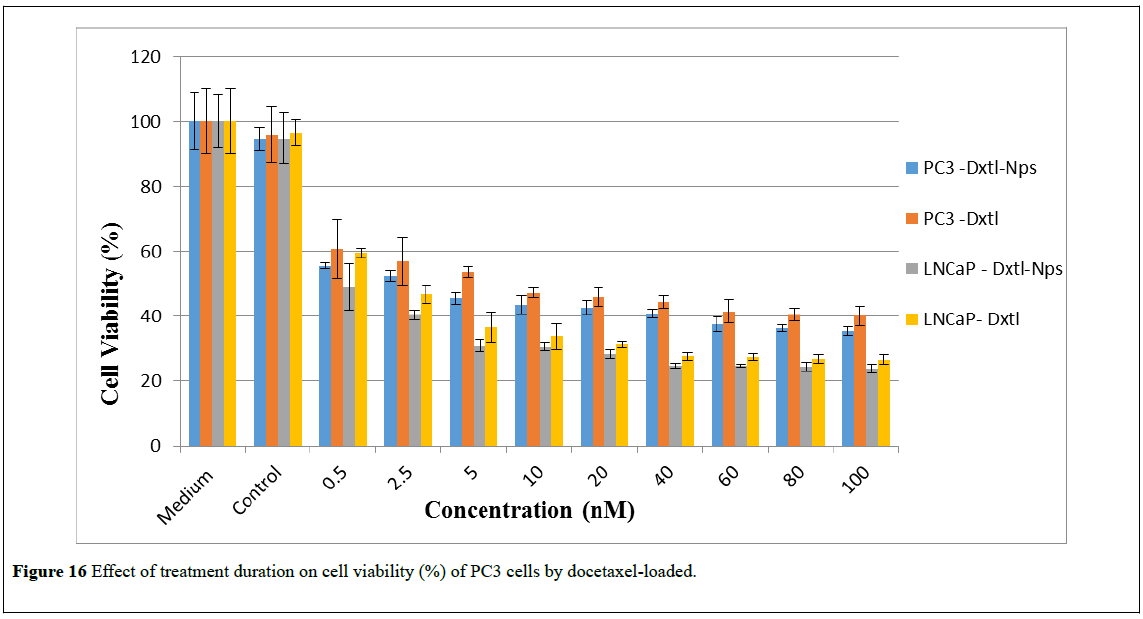
Improved cytotoxicity of docetaxel-loaded nanoparticles as compared to the free docetaxel can be explained as follows. While part of free docetaxel molecules, transported into the cytoplasm by a passive diffusion, may be transported out by p-glycoprotein (P-gp) pumps, nanoparticles are taken up by cells through an endocytosis pathway, thus resulting in a higher cellular uptake of the entrapped drug, thereby enabling them to escape from the effect of P-gp pumps [38]. Moreover, intracellular delivery of docetaxel-loaded nanoparticles allows a drug accumulation near the site of action [38,44,48]. Figures 10-15 show the effect of incubation period on the cell viability of PC3 and LNCaP cells by docetaxel-loaded nanoparticles respectively. It is evident from Figure 16 that androgen receptor sensitive-LNCaP cells are more sensitive to docetaxel than the androgen receptor insensitive-C3 cells. These results are in agreement with some previously reported cytotoxicity studies of docetaxel on LNCaP and PC3 cells, which showed that LNCaP cells are more sensitive to docetaxel than PC3 cells [44,49-52].
Figure 16: Effect of treatment duration on cell viability (%) of PC3 cells by docetaxel-loaded.
It is known that docetaxel blocks microtubule activity during cell division leading to apoptosis. It binds to the b-subunit of the tubulin heterodimer, which is the key constituent protein of cellular microtubules. The binding of docetaxel accelerates the polymerization of tubulin, and stabilizes the microtubules network, thereby inhibiting their depolymerization. This inhibition of microtubule depolymerization results in the arrest of the cell division cycle at G2/M phase, leading to apoptosis of the cancer cells eventually. Studies done by Liu, et al. [52] demonstrated that docetaxel induced apoptotic cell death in prostate cancer cells and that docetaxel treatment resulted in significant higher apoptosis in LNCaP than PC3 cells. They also demonstrated that docetaxel induced p53 phosphorylation in LNCaP cells but not in PC3 cells and that p53 status is a crucial determinant of docetaxel sensitivity in prostate cancer cells. Docetaxel treatment increases the levels of ser15 phosphorylation of p53 in a dose dependent manner in LNCaP cells, while having little effect on the levels of ser15 phosphorylation of p53 in PC3 cells [53,54]. Recently, Muenchen et. al. [54] showed that the pathway for docetaxel-induced apoptosis differs between androgen-sensitive and androgen-independent prostate cancer cells: caspase-3 and caspase-7 are cleaved in the former case, whereas caspase-8 is cleaved in the latter case.
The results clearly demonstrated that docetaxel formulated as nanoparticles was equivalent or even more effective in some cases, compared to free drug in terms of cytotoxicity against cancer cells. Both the dose exposure and the incubation time played a major role in the cell toxicity caused by docetaxel. The more marked inhibition of cell growth was obtained for longer incubation (48 hours and 72 hours) at all tested docetaxel concentrations.
Cell internalization studies
Various physicochemical properties of nanoparticles such as charge, shape, material composition, surface ligands and surface chemistry are key parameters that determine cell uptake of nanoparticles via endocytosis. Nanoparticles can be taken up by multiple endocytotic pathways, including clathrin-mediated endocytosis, caveolae mediated endocytosis, and macropinocytosis [55].
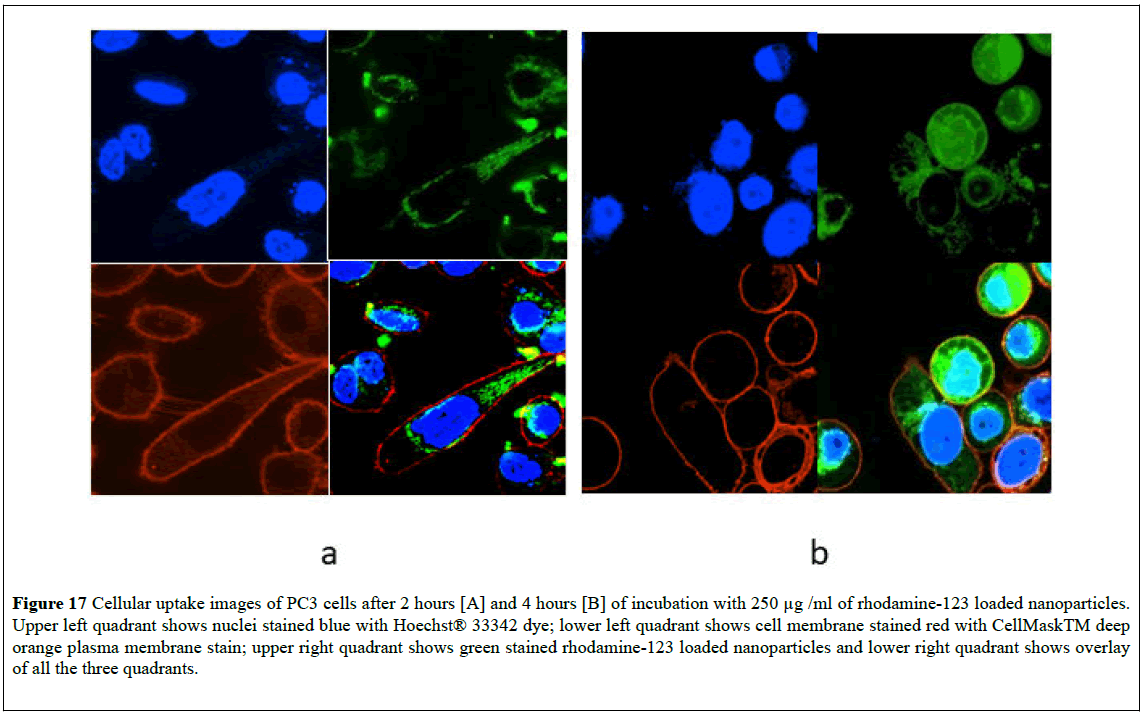
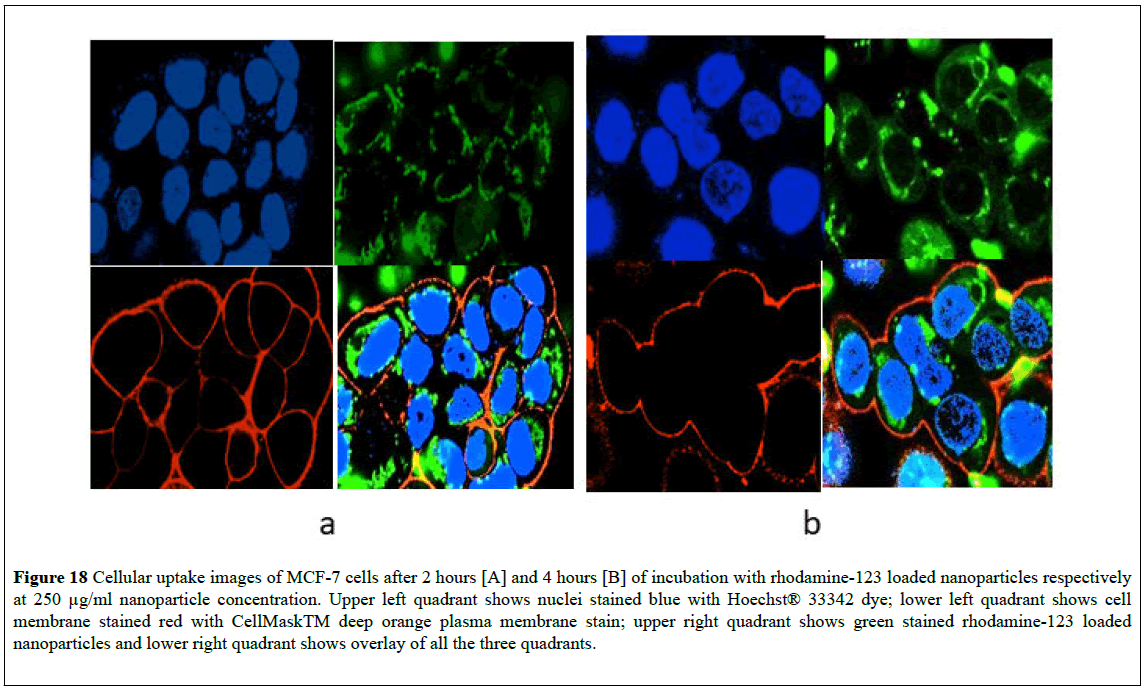
In order to study in-vitro cellular uptake of nanoparticles, rhodamine-123 loaded nanoparticles were synthesized. Figures 17 and 18 show confocal microscopic images of PC3 and MCF-7 cells respectively after exposure of the cells to rhodamine-123 labeled nanoparticles. Green fluorescence is obtained from the green channel (excitation wavelength: 488 nm, emission wavelength: 515 ± 30 nm), which represents rhodamine-123 loaded nanoparticles. The blue fluorescence is from the blue channel (excitation wavelength: 408 nm, emission wavelength: 450 ± 35 nm) which shows blue stained nuclei. The red fluorescence is from the red channel (excitation wavelength: 543, emission wavelength: 605 ± 75 nm) which shows the red stained cell membrane. The images show that the rhodamine-123-loaded nanoparticles aggregated and surrounded the nucleus and bound by the plasma membrane. The overlay images for PC3 cells (Figure 17) and MCF-7 cells (Figure 18) after 2 hours of incubation clearly show discrete nanoparticles within the cell and aggregated near the nucleus. The overlay images after 4 hours of incubation with nanoparticles for PC3 cells and MCF-7 cells show that the dye is released from the nanoparticles and is diffused within the cell around the nucleus. These results demonstrate that the pH-sensitive nanoparticles fabricated in this work can interact with the cells and can internalize within the cell to deliver the encapsulated materials to the target site.
Figure 17: Cellular uptake images of PC3 cells after 2 hours [A] and 4 hours [B] of incubation with 250 μg /ml of rhodamine-123 loaded nanoparticles. Upper left quadrant shows nuclei stained blue with Hoechst® 33342 dye; lower left quadrant shows cell membrane stained red with CellMaskTM deep orange plasma membrane stain; upper right quadrant shows green stained rhodamine-123 loaded nanoparticles and lower right quadrant shows overlay of all the three quadrants.
Figure 18: Cellular uptake images of MCF-7 cells after 2 hours [A] and 4 hours [B] of incubation with rhodamine-123 loaded nanoparticles respectively at 250 μg/ml nanoparticle concentration. Upper left quadrant shows nuclei stained blue with Hoechst® 33342 dye; lower left quadrant shows cell membrane stained red with CellMaskTM deep orange plasma membrane stain; upper right quadrant shows green stained rhodamine-123 loaded nanoparticles and lower right quadrant shows overlay of all the three quadrants.
Conclusion
We have fabricated pH-sensitive nanoparticles, by dispersion polymerization that can degrade quickly in the mildly acidic environments similar to those found in endosomes, and lysosomes, but are more stable at the physiological pH of 7.4 using a pH-sensitive bisacrylate acetal crosslinker. The release profile of docetaxel from the particles was found to be very similar to the hydrolysis profile of blank nanoparticles. This similarity shows that the release of the docetaxel occurs due to the pHdependent cleavage of the acetal crosslinker in the nanoparticle polymeric matrix. From the in-vitro cytotoxicity studies, it was evident that docetaxel formulated as nanoparticles was equivalent or more effective compared to free drug in terms of cytotoxicity against prostate cancer cells, especially after 48hours and 72 hours post treatment. Both the dose and the incubation time played a major role in the cell toxicity caused by docetaxel. The more marked inhibition of cell growth was obtained for longer incubation (48 hours and 72 hours) at all tested docetaxel concentrations. Furthermore, LNCaP cells appeared to be more sensitive to docetaxel compared to PC3 cells. Also, in-vitro cytotoxicity assay showed that the blank nanoparticles are biocompatible with no toxicity for the duration of the assay comparable to medium-only treated controls. From the cell internalization experiment, it was evident that the nanoparticles were internalized into the cells in less than 2 hours probably by nonspecific endocytosis. These novel pH-sensitive nanoparticles would not only offer several advantages over conventional drug therapies but also are expected to overcome side effects related to dosing and toxicity of docetaxel. Thus, these nanoparticles are suitable as pH-responsive drug delivery system for the treatment of cancer and other diseases that need targeting to the biophase.
Conflict of Interest
The authors confirm that the content of this research article has no conflict of interest.
Acknowledgements
Research reported in this publication was supported by NCI/NIH Grant #: NCI/NIH Grant #: 1SC1CA199810-01. This work was carried out in facilities supported by NCRR/NIH Grants #1 C06 RR 020608-01 and #1 C06 RR 14469-01.
REFERENCES
- Luo Y, Ling Y, Pang J, et al. Docetaxel loaded oleic acid-coated hydroxyapatite nanoparticles enhance the docetaxel-induced apoptosis through activation of caspase-2 in androgen independent prostate cancer cells. J Control Rel. 2010;147:278-88.
- Kornek GV, Haider K, Kwasny W, et al. Treatment of advanced breast cancer with docetaxel and gemcitabine with and without human granulocyte colony-stimulating factor. Clinical Cancer Research 2002;8:1051-6.
- Liu B, Yang M, Li X, et al. Enhanced efficiency of thermally targeted taxanes delivery in a human xenograft model of gastric cancer. J Pharm Sci. 2008;97:3170-81.
- Sanna V, Roggio AM, Posadino AM, et al. Novel docetaxel-loaded nanoparticles based on poly(lactide-co-caprolactone) and poly(lactide-co-glycolide-co-caprolactone) for prostate cancer treatment: formulation, characterization, and in vitro cytotoxicity studies. Nanoscale Res Let. 2011;6:1-9.
- Zhao P, Astruc D. Docetaxel nanotechnology in anticancer therapy. Chem Med Chem. 2012;7:952-72.
- Gulloti E, Yeo Y. Extracellularly activated nanocarriers: A new paradigm of tumor targeted drug delivery. Mol Pharm. 2009;6:1041-51.
- Du JZ, Du XJ, Mao CQ, et al. Tailor-Made Dual pH-Sensitive Polymer-Doxorubicin Nanoparticles for Efficient Anticancer Drug Delivery. J Am Chem Soc. 2011;133:17560-63.
- Oku N, Tokudome Y, Asai T, et al. Evaluation of Drug Targeting Strategies and Liposomal Trafficking. Cur Pharm Des. 2000;6:1669-91.
- Gillies ER, Goodwin AP, Frechet JM. Acetals as pH-sensitive linkages for drug delivery. Bioconjugate Chem. 2004;15:1254-63.
- Gillies ER, Frechet JM. pH-responsive copolymer assemblies for controlled release of doxorubicin. Bioconjugate Chem. 2005;16:361-68.
- Akala EO, Kopeckova P, Kopecek J. Novel pH Sensitive Hydrogels with Adjustable Kinetics of Swelling. Biomat. 1998;19:1037-47.
- Gilles ER, Frechet JM. Development of acid-sensitive copolymer micelles for drug delivery. Pure App Chem. 2004;76:1295-1307.
- Bulmus V, Chan Y, Nguyen Q, et al. Synthesis and characterization of degradable p(HEMA) microgels: Use of acid-labile crosslinker. Macromol Biosci. 2007;7:446-55.
- Martinez FC, Carriona B, Lucio MI, et al. Enhanced docetaxel-mediated cytotoxicity in human prostate cancer cells through knockdown of cofilin-1 by carbon nanohorn delivered siRNA. Biomat. 2012;33:8152-9.
- Chan Y, Bulmus V, Zareie MH, et al. Acid-cleavable polymeric core-shell particles for delivery of hydrophobic drugs. J Contr Rel. 2006;115:197-207.
- Vitullo VP, Pollack R, Faith WC, et al. Substituent effects in acetal hydrolysis. J Am Chem Soc. 1974;96:6682-85.
- Puri R, Berhe SA, Akala EO. pH-Sensitive polymeric nanoparticles fabricated by dispersion polymerization for the delivery of bioactive agents. Pharma Nanotech. 2017;5:1-28.
- Bharali D, Khalil M, Simone TM, et al. Nanoparticles and cancer therapy: A concise review with emphasis on dendrimers. Int J Nanomed. 2009;4:1-7.
- Ogunwuyi O, Adesina S, Akala EO. D-Optimal mixture experimental design for stealth biodegradable crosslinked docetaxel-loaded poly-caprolactone nanoparticles manufactured by dispersion polymerization. Pharmazie 2015;70:165-76.
- Adesina SK, Wight SA, Akala EO. Optimization of the fabrication of novel stealth PLA based nanoparticles by dispersion polymerization using D-optimal mixture design. Drug Dev Ind Pharm. 2014;40:1547-56.
- Akala EO, Okunola O. Novel stealth degradable nanoparticles prepared by dispersion polymerization for the delivery of bioactive agents. Part I. Pharm Ind. 2013;75: 1191–1196; Part II Pharm Ind. 2013;75:1346-52.
- Ogunwuyi O, Kumari N, Smith KA, et al. Antiretroviral Drugs-Loaded Nanoparticles Fabricated by Dispersion Polymerization with Potential for HIV/AIDS Treatment. Infectious Diseases: Research and Treatment 2016;9:21-32.
- Adesina SK, Holly A, Kramer-Marek G, et al. Polylactide-based Paclitaxel-loaded Nanoparticles Fabricated by Dispersion Polymerization: Characterization, Evaluation in Cancer Cell Lines, and Preliminary Biodistribution Studies. J Pharma Sci. 2014;103:2546-55.
- Smith KA, Lin X, Bolshakov O, et al. Activation of HIV-1 with Nanoparticle-Packaged Small Molecule Protein Phosphatase-1-Targeting Compound Sci Pharm. 1502-01.
- Gillies ER, Frechet JM. pH-responsive copolymer assemblies for controlled release of doxorubicin. Bioconjugate Chem. 2005;16:361-68.
- Chan Y, Wong T, Byrne F. et al. Acid-labile core cross-linked micelles for pH triggered release of antitumor drugs. Biomacromol. 2008;9:1826-36.
- Bulmus V, Chan Y, Nguyen Q, et al. Synthesis and characterization of degradable p(HEMA) microgels: Use of acid-labile crosslinker. Macromol Biosci. 2007;7:446-55.
- Win KY, Feng SS. Effects of particle size and surface coating on cellular uptake of polymeric nanoparticles for oral delivery of anticancer drugs. Biomat. 2005;26:2713-22.
- Ferlini C, Biselli R, Nisini, R, et al. Rhodamine-123: A useful probe for monitoring T-cell activation. Cytomet. 1995;21:284-93.
- Murthy N, Thong YX, Schuck S, et al. A novel strategy for encapsulation and release of proteins: Hydrogels and microgels with acid-labile acetal crosslinkers. J Am Chem Soc. 2002;124:12398-99.
- Yin W, Akala EO, Taylor R.E. Design of naltrexone-loaded hydrolysable crosslinked nanoparticles. Int J Pharm. 2002;244:9-19.
- Tao W, Zeng X, Liu T, et al. Docetaxel-loaded nanoparticles based on star-shaped mannitol-core PLGA-TPGS diblock copolymer for breast cancer therapy. Acta Biomaterialia 2013;9:8910-20.
- Yanasarn N, Sloat BR, Cui Z. Nanoparticles engineered from lecithin-in-water emulsions as a potential delivery system for docetaxel. Int J Pharmaceutics, 2009;379:174-180.
- Ma Y, Zheng Y, Zeng X, et al. Novel docetaxel-loaded nanoparticles based on PCL-Tween 80 copolymer for cancer treatment. Int J Nanomed. 2011;6: 2679-88.
- Mateja C, Janko K, Julijana K. Cystatin incorporated in poly(lactide-co-glycolide) nanoparticles: development and fundamental studies on preservation of its activity. Euro J Pharma Sci. 2004;22:357-64.
- Lundberg BBPreparation and characterization of polymeric pH-sensitive stealth® nanoparticles for tumor delivery of a lipophilic prodrug of paclitaxel. International J Pharma. 2011;408:208-12.
- Chaouhan R, Bajpai AK. Real time in vitro studies of doxorubicin release from PHEMA nanoparticles. J Nanobiotech. 2009;7:1-12.
- Mei L, Zhang Y, Zheng Y, et al. A Novel docetaxel-loaded Poly (ε-Caprolactone)/Pluronic F68 nanoparticle overcoming multidrug resistance for breast cancer treatment. Nanoscale Research Letters 2009;4:1530-39.
- Dong Y, Feng SS. Methoxy poly (ethylene glycol)-poly(lactide) (MPEG-PLA) nanoparticles for controlled delivery of anticancer drugs. Biomat. 2004;25:2843-9.
- Zheng D, Li X, Xu H, et al. Study on docetaxel-loaded nanoparticles with high antitumor efficacy against malignant melanoma. Acta Biochimica et Biophysica Sinica 2009; 41:578-87.
- Zhang L, Yang M, Li Y, et al. 10- hydroxycamptothecin loaded nanoparticles: Preparation and antitumor activity in mice. J of Contr Rel. 2007;119:153-62.
- Liu Y, Li K, Liu B, et al. A strategy for precision engineering of nanoparticles of biodegradable copolymers for quantitative control of targeted drug delivery. Biomat. 2010;31:9145-55.
- Zubris KAV. Polymeric nanoparticles for the intracellular delivery of paclitaxel in lung and breast cancers. (Doctoral Dissertation), Retrieved from ProQuest Dissertations and Theses 2011.
- Luo Y, Ling Y, Pang J, et al. Docetaxel loaded oleic acid-coated hydroxyapatite nanoparticles enhance the docetaxel-induced apoptosis through activation of caspase-2 in androgen independent prostate cancer cells. J Contr Rel. 2010;147:278-88.
- Fabbril F, Brigliadoril G, Carlonil S, et al. Zoledronic acid increases docetaxel cytotoxicity through pMEK and Mcl-1 inhibition in a hormone-sensitive prostate carcinoma cell line. J Transl Med. 2008;6:1-10.
- Giannakakou P, Robey R, Fojo T, et al. Low concentrations of paclitaxel induce cell type-dependent p53, p21 and G1/G2 arrest instead of mitotic arrest: Molecular determinants of paclitaxel induced cytotoxicity. Oncogene 2001;20:3806-13.
- Liebmann JE, Cook JA, Lipschultz C, et al. Cytotoxic studies of paclitaxel (Taxol®) in human tumour cell lines. Br J Cancer 1993;68:1104-09.
- Ryckewaerta DL, Carpentiera R, Lipkaa E, et al. Development of innovative paclitaxel-loaded small PLGA nanoparticles: Study of their antiproliferative activity and their molecular interactions on prostatic cancer cells. Int J Pharma. 2013;454:712-19.
- Li Y, Li X, Hussainy M, et al. Regulation of microtubules, apoptosis, and cell cycle-related genes by Taxotere in prostate cancer cells analyzed by microarray. Neoplasia,2004;6:158-67.
- Wiltshire C, Singh BL, Stockley J, et al. Docetaxel-Resistant Prostate Cancer Cells Remain Sensitive to S-Trityl-L-Cysteine–Mediated Eg5 Inhibition. Mol Cancer Therap. 2010;9:1730-9.
- Szliszka E, Bronikowska J, Majcher A, et al. Enhanced sensitivity of hormone-refractory prostate cancer cells to tumour necrosis factor-related apoptosis-inducing ligand (TRAIL) mediated cytotoxicity by taxanes. Cent Euro J Urology 2009;62:29-34.
- Liu C, Zhu Y, Lou W, et al. Functional p53 determines docetaxel senisitivity in prostate cancer cells. Prostate 2013;73:418-27.
- Conagin A, Barbin D. Bonferroni’s and Sidak’s modified tests. Scientia Agricola 2006;63:70-6.
- Muenchen HJ, Poncza PJ, Pienta KJ. Different docetaxel-induced apoptotic pathways are present in prostate cancer cell lines LNCaP and PC-3. Urology 2001;57: 366-70.
- Zubris KAV. Polymeric nanoparticles for the intracellular delivery of paclitaxel in lung and breast cancers. (2011 Doctoral Dissertation), Retrieved from ProQuest Dissertations and Theses.