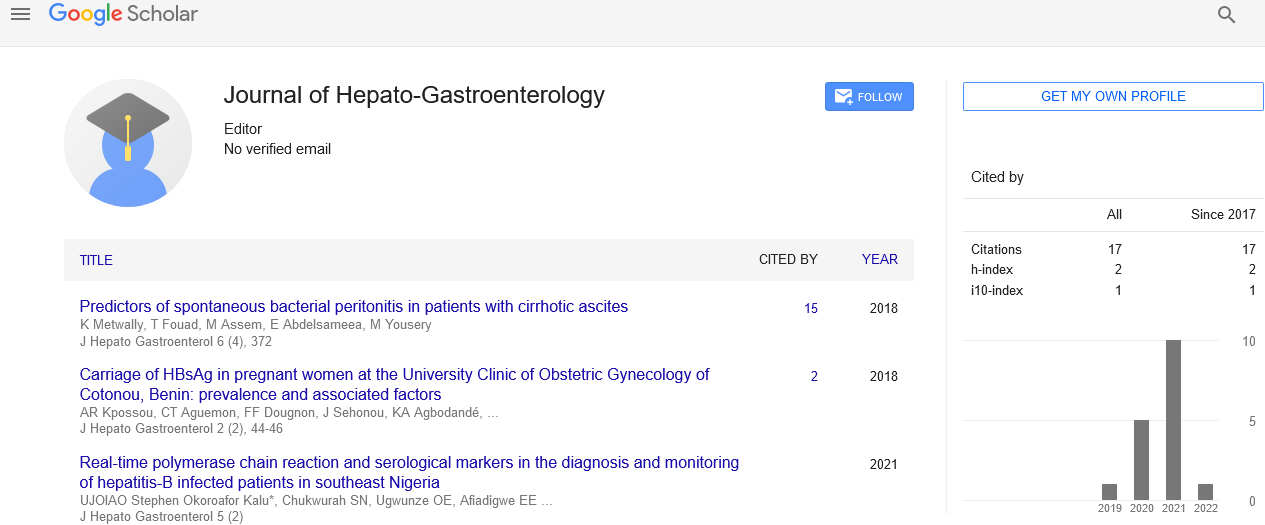
AhMITE1 influences the expression of trait-specific genes in peanut ( Arachis hypogaea L.)
2 Department of Frontier Research and Development, Kazusa DNA Research Institute, Chiba, Japan
Received: 04-Dec-2022, Manuscript No. PULHG-23-5831; Editor assigned: 06-Dec-2022, Pre QC No. PULHG-23-5831 (PQ); Reviewed: 20-Dec-2022 QC No. PULHG-23-5831; Revised: 20-Jan-2023, Manuscript No. PULHG-23-5831 (R); Published: 27-Jan-2023
Citation: Bhat RS, Choudhari R, Jadhav MP, et al. AhMITE1 influences the expression of trait-specific genes in peanut (Arachis hypogaea L.). Clin Cardiol J 2023;7(1):1-2.
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
Arachis hypogaea Miniature Inverted repeat Transposable Element 1 (AhMITE1 ), a class II non-autonomous transposable element, was analyzed for its influence on the rate of expression of 25 trait specific genes in peanut. Of them, 14 genes could be considered for expression analysis based on the primer efficiency and availability of the minimal template transcripts in the samples for qRT-PCR. The allele with AhMITE1 insertion showed similar frequencies of up regulation and down regulation across 11 genotypes on 21st day after sowing. Overall, the region (upstream, UTR, intron, downstream etc.) of AhMITE1 insertion did not influence gene regulation, but the study revealed the importance of AhMITE1 in influencing the gene regulation and thereby the phenotype in peanut.
Keywords
Peanut; AhMITE1; AhTE markers; Trait specific genes; Gene regulation
Introduction
Peanut (Arachis hypogaea L.) genome has a large portion (~74%) consisting of Transposable Elements (TEs). This repetitive genome probably contributes for extending the diversity and accelerating the rate of genome evolution by altering gene expression patterns and phenotypes so that peanut can adapt more rapidly to new environmental conditions. Considering the useful properties of the TEs such as their large number in the genome, high rate of insertion polymorphism, transposition preference for genic region and ease of detection, a new class of markers (Arachis hypogaea TE; AhTE) were proposed and later 3,996 markers were developed using transposon display, in silico analysis and whole genome sequence search. They were mapped to identify the genomic regions and the candidate genes for the taxonomic, morphological and the productivity traits. Some of trait specific markers were also genic since the AhMITE1 insertion/excision site corresponded to the genic region [1-3].
Literature Review
Since the transposable elements influence the phenotype by altering the gene expression by down regulation or up regulation, it is important to analyze the expression levels of trait specific genes among the diverse genotypes of peanut to study the influence of AhMITE1 insertion on gene regulation because such reports are limited in peanut. Therefore, an effort was made in this study to check the allelic variation at the genic AhTE marker loci linked to important traits using the diverse genotypes of peanut. Further, the expression levels of the corresponding genes were assessed using qRT-PCR for ascertaining the influence of AhMITE1 insertion on gene regulation and its plausible influence on the phenotype in peanut [4-7].
Eighteen diverse genotypes of peanut were used for allele profiling at 25 AhTE marker loci of the 18, only 11 genotypes were used for qRT-PCR of the 25 AhMITE1 associated trait specific genes along with a reference gene (Glucose 6-Phosphate 1-Dehydrogenase (G6PD); Arahy. XC1VLW on chromosome 1/Arahy. 74FNJK on chromosome 11). The seeds were sown in pots, which were placed in the greenhouse. Care was taken to grow healthy plants. Leaf samples were collected on 21 Days after Sowing (DAS) from all the genotypes. Total RNA was isolated using Qiagen Rneasy mini kit (Qiagen, USA) as per manufacturer’s protocol. RNA was treated with DNase using column DNase Treatment in Qiagen Rneasy mini column as per manufacturer’s instructions. RNA was quantified using a Nano Drop spectrophotometer (ND-2000, thermo fisher scientific, USA). Total RNA integrity was analyzed with an Agilent 2100 Bio analyzer (Agilent technologies, Palo Alto, CA) according to the manufacturer’s instructions. The 18S and 28S rRNA ratio was obtained from 2100 expert software (Agilent technologies) and the RNA integrity number was obtained from RIN beta version software (Agilent technologies) [8].
The primers were designed for the selected genes using primer3 plus considering exotic and coding region of the transcripts. Total RNA was converted to cDNA using Affinity Script qPCR cDNA synthesis kit (Agilent technologies, USA) as per manufacturer`s protocol. Primer efficiency was checked using the original concentration (25 ng/μL) and three dilutions (1:4, 1:16 and 1:64). Based on the R2 threshold of 0.95, the primer efficiency was calculated. The primers were validated for their specificity using pooled cDNA from all the samples. The expression levels of the selected genes were analyzed using SYBR green chemistry (Brilliant II SYBR green qPCR master mix (Agilent technologies, USA) using 1 μl (40 ng/μl) of cDNA with technical replicates. The assay was carried out in strata gene mx3005P instrument (Agilent technologies, USA). The mean Cycle Threshold (CT) value of technical replicates was used to calculate the relative expression levels of the genes. The relative quantification of genes was analyzed using the standard 2^-ΔΔCt as described by using G6PD as the reference gene to normalize the qPCR experiment. The fold change across the samples were compared using the t test [9,10].
Peanut genotypes when checked for the allelic variation at the 25 AhTE genic and trait specific marker loci, showed two types of alleles “A” type with AhMITE1 insertion, and “B” type without AhMITE1 insertion at given marker locus. The frequency of “B” allele (0.76) was more than that of “A” allele (0.24) across 25 AhTE marker loci [11].
Discussion
RNA isolated from the leaves of 21 days old seedlings of the eleven genotypes were optimal with respect to the purity, concentration and integrity. Primer efficiency of ≥ 90% with an R2 value ≥ 0.95 was observed for 17 genes (excluding the reference gene), and they were considered for the expression analysis. Of them, 14 genes showed Ct value less than 28 with the cDNA pooled from the eleven samples, indicating the availability of optimum cDNA templates for amplification. For the remaining three genes (Araip.Q2YHX, Araip. AP706 and Araip.ISU30), poor amplification was observed. Non-template control samples (without the template cDNA) showed either no amplification or amplification with very high mean Ct values. Specific amplification was verified by melting curve analysis, where a single peak was observed for all the 14 genes, while the non-template control samples did not show any peak, except for Aradu. E02FY, which could be due to the primer dimers. Specific amplification was also confirmed through gel electrophoresis. The reference gene also showed the specific amplification with a Ct value of 26.66 [12].
When the mean Ct values of the “A” and “B” alleles at 14 genes were compared across 11 genotypes, the A allele showed down regulation at seven genes, and up regulation at the remaining seven genes. Previously, similar frequencies of up and down regulated expression changes were observed by interestingly, the gene Aradu. BZ12G corresponding to the marker AhTE0233 associated with resistance to LLS and rust, showed significant up regulation in the “A” allele (found in the susceptible genotypes) over the “B” allele (observed in resistant genotypes). A allele had AhMITE1 insertion at 1,399 bp (fourth exon) of Aradu. BZ12G located on A01 chromosome, and this gene was found to code for sterol C4-methyl oxidase, which plays a key role in fatty acid biosynthetic process [13-16].
In order to obtain a precise understanding on the influence of AhMITE1 on gene expression, TMV 2 and its EMS-derived mutant TMV 2-NLM were checked for the expression of the trait specific genes since these genotypes provide an opportunity to reduce the noise due to background mutations. Again, the “A” allele showed down regulation at 10 genes, and up regulation at three genes. In general, the gene regulation was not influenced by the region (upstream, UTR, intron, downstream etc.) of insertion of AhMITE1. The previous report of down regulation of an intron less gene (AhFAD2B coding for microsomal oleoyl-PC desaturase) in peanut was associated with AhMITE1 insertion in the coding region. Though down regulation of the genes is more common with the insertion of TEs a few reports indicate up regulation similar to the gene Aradu. BZ12G and Aradu. 4RE2S in this study [17-20].
Conclusion
Overall, TEs influence the gene regulation through diverse mechanisms insertions into exons usually lead to loss of function or alteration in the gene product, 2) insertion into introns can cause new alternative splicing and 3) insertion into upstream or downstream regulatory sequences may lead to either up regulation or down regulation, depending on the role of a particular regulatory sequence. TEs can induce formation of heterochromatic islands, through various epigenetic silencing mechanisms which may lead to down regulation of genes. Therefore, a better understanding of gene expression and function as influenced by AhMITE1 could help in gene based innovative breeding methods in peanut in future.
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- Bertioli DJ, Jenkins J, Clevenger J, et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat Genet. 2019; 51(5):877-84.
[Crossref] [Google Scholar] [PubMed]
- Bhat RS, Patil VU, Chandrashekar TM, et al. Recovering flanking sequence tags of a miniature inverted repeat transposable element by thermal asymmetric interlaced-PCR in peanut. Curr Sci. 2008;95(4):452-3.
- Dubin MJ, Scheid OM, Becker C, et al. Transposons: A blessing curse. Curr Opin Plant Biol. 2018;42:23-9.
[Crossref] [Google Scholar] [PubMed]
- Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331-68.
[Crossref] [Google Sholar] [PubMed]
- Gayathri M, Shirasawa K, Varshney RK, et al. Development of AhMITE1 markers through genome wide analysis in peanut (Arachis hypogaea L.). BMC Res. 2018;11(1):1-6.
[Crossref] [Google Scholar] [PubMed]
- Gowda MV, Bhat RS, Motagi BN, et al. Association of high frequency origin of late leaf spot resistant mutants with AhMITE1 transposition in peanut. Plant Breed. 2010;129(5):567-9.
- Hake AA, Shirasawa K, Yadawad A, et al. Mapping of important taxonomic and productivity traits using genic and non-genic transposable element markers in peanut (Arachis hypogaea L). PLoS one. 2017;12(10): 0186113.
[Crossref] [Google Sholar] [PubMed]
- Kuang H, Padmanabhan C, Li F, et al. Identification of Miniature Inverted repeat Transposable Elements (MITEs) and biogenesis of their siRNAs in the Solanaceae: New functional implications for MITEs. Genome Res. 2009;19(1):42-56.
[Google Scholar] [PubMed]
- Lu C, Chen J, Zhang Y, et al. Miniature Inverted repeat Transposable Elements (MITEs) have been accumulated through amplification bursts and play important roles in gene expression and species diversity in Oryza sativa. Mol Biol Evol. 2012;29(3):1005-17.
- Makarevitch I, Waters AJ, West PT, et al. Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS genetics. 2015;11(1):1004915.
- Mao H, Wang H, Liu S, et al. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat Commun. 2015;6(1):8326.
[Crossref] [Google Scholar] [PubMed]
- Naito K, Zhang F, Tsukiyama T, et al. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature. 2009;461(7267):1130-4.
[Crossref] [Google Scholar] [PubMed]
- Patel M, Jung S, Moore K, et al. High oleate peanut mutants result from a MITE insertion into the FAD2 gene. Theor Appl Genet. 2004; 108:1492-502.
[Crossref] [Google Scholar] [PubMed]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29(9):45.
[Crossref] [Googlesholar]
- Shen J, Liu J, Xie K, et al. Translational repression by a miniature inverted-repeat transposable element in the 3′ untranslated region. Nat Commun. 2017;8(1):14651.
[Crossref] [Google Scholar] [PubMed]
- Shirasawa K, Hirakawa H, Tabata S, et al. Characterization of active miniature inverted-repeat transposable elements in the peanut genome. Theor Appl Genet. 2012;124:1429-38.
- Shirasawa K, Koilkonda P, Aoki K, et al. In silico polymorphism analysis for the development of simple sequence repeat and transposon markers and construction of linkage map in cultivated peanut. BMC Plant Biol. 2012;12:1-3.
[Crossref] [Google Scholar] [PubMed]
- Wessler SR. Transposable elements associated with normal plant genes. Physiol Plant. 1998;103(4):581-6.
- Yang G, Lee YH, Jiang Y, et al. A two edged role for the transposable element Kiddo in the rice ubiquitin2 promoter. The Plant Cell. 2005; 17(5):1559-68.
[Crossref] [Google Scholar] [PubMed]
- Yasuda K, Ito M, Sugita T, et al. Utilization of transposable element mPing as a novel genetic tool for modification of the stress response in rice. Mol Plant Breed. 2013;32:505-16.
[Crossref] [Google Scholar] [PubMed]