High-resolution melting curve analysis for rapid detection of Isoniazid-resistant Mycobacterium tuberculosis in Qinghai Province
2 Qinghai Center for Disease Control and Prevention, Xining, China
Received: 15-Oct-2018 Accepted Date: Nov 03, 2018; Published: 13-Nov-2018
Citation: Zhang Y, Wang Z, Li B, et al. High-resolution melting curve analysis for rapid detection of Isoniazid-resistant Mycobacterium tuberculosis in Qinghai Province. J Microbiol Biotechnol Rep. 2018;2(3): 50-53.
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
Objectives: To evaluate the diagnostic value of high-resolution melting curve (HRM) analysis to isoniazid-resistant Mycobacterium tuberculosis (MTB) derived from patients on the Qinghai-Tibet Plateau, compared with DNA sequencing. Methods: A total of 96 clinical isoniazid (INH) resistant isolates, determined by drugs susceptibility testing using the proportion method on Lowenstein-Jensen (L-J) medium, were included into analysis. Codon KatG315 of these isolates was analysed by both HRM and DNA sequencing, and results were compared. Results: Of the 96 INH resistant strains, 55 isolates with katG315 mutations were detected by HRM, accounting for 57.3%, and 50 strains (52.1%) were found by sequencing, which is considered as gold standard for DNA mutations. The sensitivity of HRM is 88.0%, the specificity is 76.1%, Mc-Nemar test P=0.332 and kappa coefficient is 0.644. Conclusion: HRM and sequencing have moderate consistency in detecting KatG315 mutations. Furthermore, HRM can detect heterogeneous resistance. It would be a superior method for the detection of INH resistance-related mutations at basic level units of Qinghai province.
Keywords
Mycobacterium tuberculosis; High-resolution melting curve analysis; Isoniazid-resistant; KatG
Introduction
Early antibiotic treatment is crucial for tuberculosis (TB) treatment. The multidrug resistant tuberculosis (MDR-TB) and extensively drug resistant tuberculosis (XDR-TB), mainly resulting from incorrect treatment, have severely blocked TB control worldwide. According to WHO global tuberculosis report, in 2016, there were 10.4 million people infected with TB, 1.7 million patients died of TB- related diseases and 490 thousand new cases suffered from MDR-TB [1]. It was estimated that more than 400 thousand TB patients turned into MDR due to inappropriate monitoring of drug-resistant TB every year [2]. The L-J proportion method is the gold standard for the diagnosis of drugresistant TB with long detective periods (more than 2 months). However, It is essential to detect drug resistance rapidly, not only for the guidance of initial treatment, but also for effective drug-resistant TB control [3-5]. Therefore, the development of rapid detection methods is the urgent priority. There have been several molecular biological technologies detecting resistance related gene mutations in order to shorten the time of diagnosis. However, most of the molecular biological means are expensive, not-rapid enough and hard to perform. High-resolution melting analysis (HRM) which based on the physical properties of nucleic acid, utilizes the saturated DNA dye to describe small distinctions in DNA sequence by generating discrepant melting curve. Also, it is not limited by mutation sites or types, and has been successfully applied to mutation scanning, pre-sequencing screening, SNP and methylation detection [6,7]. It is a rapid, accurate, simple and economical molecular technology for the detection of mutated genes [8]. INH is a commonly used drug for treating TB and is activated by KatGencoded catalase peroxidase [9]. Codon katG315 is the major mutation site of INH resistance, which accounts for about 50-90% of INH resistant strains [5,10]. Studies reveal that the mutation frequency of katG is 100 times higher than that of rifampin resistant gene rpoB, and KatG mutation is more likely to produce other related genes’ mutation. It is considered to be the first step for the evolution of MDR-TB [5]. Therefore, investigating katG315 is of great significance for clinical therapy. Qinghai province lies in the north-east of Qinghai-Tibet Plateau, with an average altitude of more than 3000 meters and harsh natural environment, and the medical and health services here are lagging behind the other parts of country. TB prevails widespread in Qinghai, and TB prevalence is higher than that of many other areas in China [11]. This study aimed to explore HRM analysis for the detection of drug-resistance MTB, thus to provide a rapid and cost-effective manner at grass-roots units of Qinghai province.
Materials and Methods
Clinical isolates of MTB strains
Total 96 INH resistant clinical MTB strains were provided by Qinghai Center for Disease Control and Prevention (CDC), all isolates were collected from hospitals or CDC in 7 cities of Qinghai Province, and the standard strain H37Rv was provided by Chinese CDC.
Drugs susceptibility testing (DST)
DST were performed on L-J medium containing 0.2 μg/ml INH as described previously [12].
DNA preparation
DNAs of MTB were extracted by boiling: isolates were inactivated at 85ºC for 1 h, then boiled for 30 min. After centrifugation at 12,000 rpm for 5 min, the supernatants were collected as DNA samples. The concentration of DNA samples were adjusted to 5-30 ng/μl by NanoDrop 2000, which is the best range for HRM [13].
Detection of KatG gene by HRM
The primers were designed to generate short PCR products, which fit HRM analysis better. The sequences of HRM primers are shown in Table 1. Programs were performed on LightCycler96 (Roche). A reaction mixture contained 5 μl of HRM Analysis PreMix, 1 μl of each prime (1 umol/L), 1 μl of DNA and 2 μl of ddH2O. The amplification was performed using the conditions listed in Table 2. After the first HRM, for the samples showing as a single-peak curve, a second HRM was performed—the PCR products of H37Rv and first PCR products of samples were mixed with equal volumes, then a second round PCR was conducted in a shortened version: 1 cycle of 95ºC for 5 min, 5 cycles of 95ºC for 10 s, 60ºC for 10 s, 72ºC for 10s. This step can generate DNA hybridized double strands between samples and H37Rv, then HRM analysis was used again as described in Table 3. It was expected to form double or blunt peak for mutated strains, which showed as single peak curve in the first HRM.
Table 1: Primers for PCR of sequencing and HRM
| Primer | Products length | |
|---|---|---|
| PCR for HRM | F:TCGTATGGCACCGGAACC R:CAGCTCCCACTCGTAGCC |
123bp |
| PCR for Sequencing | F:GATCGTCGGCGGTCACACTT R:CGTTGACCTCCCACCCGACT |
731bp |
Table 2: High-resolution melting curve analysis
| Program | Temp(°C) | Time(s) | Cycles | Acquisition(per°C) |
|---|---|---|---|---|
| Pre-denaturation | 94 | 300 | 1 | _ |
| denaturation | 94 | 10 | _ | |
| annealing | 60 | 10 | 25 | _ |
| extension | 72 | 10 | _ | |
| HRM | 94 40 70-94 |
60 60 _ |
1 | _ _ 25 |
Table 3: The cross table for the analysis of Sequencing and HRM
| HRM | Sequencing | |
|---|---|---|
| Mutation | No-mutation | |
| Mutation | 44 | 11 |
| No-mutation | 6 | 35 |
Note: P=0.332>0.05
KatG gene sequencing
KatG gene of 96 Isoniazid resistant strains were amplified by PCR, the primers are shown in Table 1. The PCR products were visualized by 2% agarose gel electrophoresis and then were sent for sequencing. H37Rv was included as positive control and sterile distilled water as negative control.
Statistical analysis
SPSS18.0 was used for data analysis, DNA sequencing was used as the reference pattern. The kappa coefficient was used to analyze the consistency of these two results. Kappa ≥ 0.75 was considered highly consistent, 0.4 ≤ kappa<0.75 was moderately consistent, and kappa<0.4 was poorly consistent. Mc-Nemar test was also performed between these two methods, P<0.05 indicating the difference between these two methods was statistically significant.
Results
Results of HRM
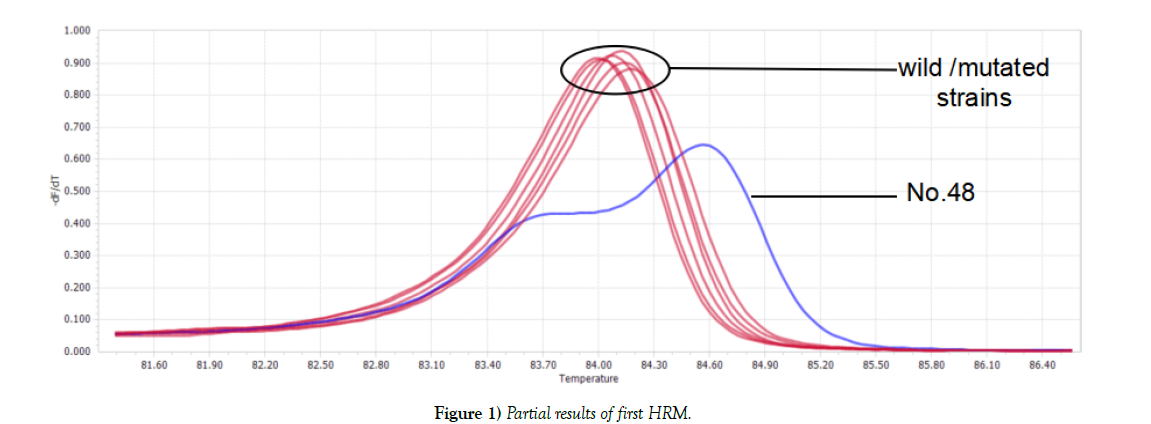
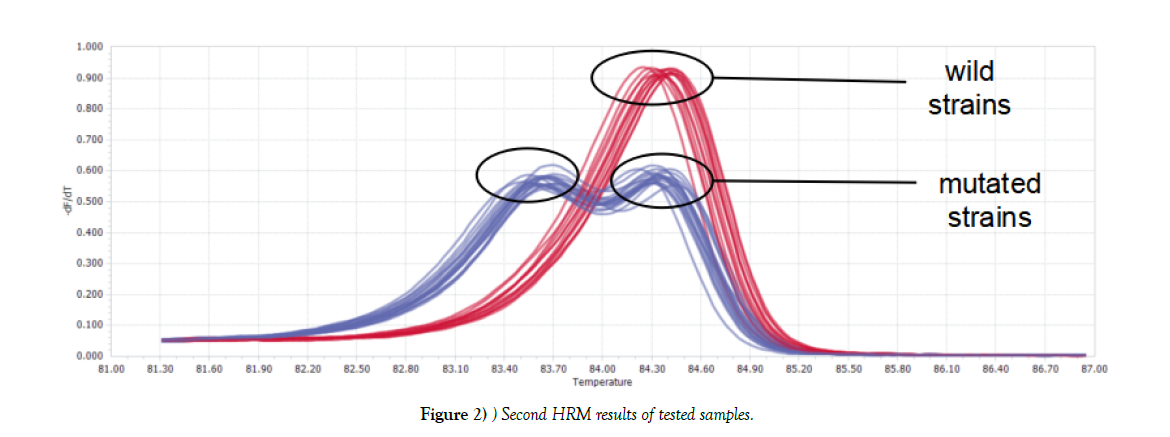
Two kinds of graphs generated after first HRM: one was double-peak curve, indicating there were two different types of genes in one sample (strain NO.48). The other (the rest tested strains) was single peak curve meaning the sample contained only one kind of DNA, which considered as wild type or mutated type. Parts of results were shown in Figure 1. For Single peak samples, second HRM need to be conducted. Second HRM also showed two types of curves: the single peak curve representing the sample was wild isolate, while the double-peak curve expressing mutated strain (Figure 2).
Sequencing results of Katg Gene
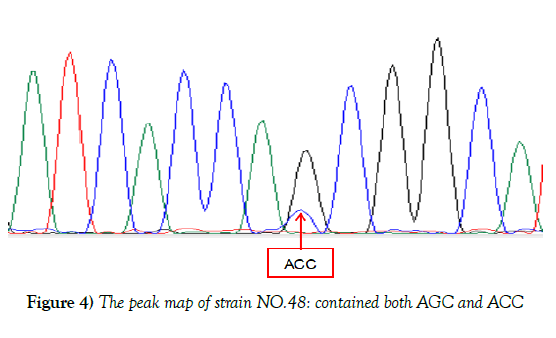
In reference to the KatG gene of H37Rv, 50 strains were found mutated at katG315 accounting for 52.1%, 2 types of mutations were observed, 49 isolates mutated from AGC to ACC, one was AAC, partial results were shown in Figure 3. Although sequencing result of NO.48 showed no mutation, the peak map revealed that there were two mutations at KatG315 (AGC and ACC), which was consistent with the result of HRM curve, suggesting that strain 48 might be heterogeneous drug-resistant bacteria, shown in Figure 4. Of the 96 resistant isolates, 50 were mutated strains in code KatG315 detected by sequencing, 55 were found by HRM, 11 of which were not found mutations by sequencing, and 6 strains were found mutations by sequencing but not HRM. Sequencing was taken as the gold standard, the sensitivity of HRM for mutations detection was 88%, the specificity was 76.1%, the positive predictive value was 80%, and the negative predictive value was 85.4%. The diagnostic accuracy rate was 82.3%, and Youden’s index was 0.641. Difference between Sequencing and HRM was not statistically significant according to McNemar test (P=0.332). The kappa coefficient was 0.644, indicating the results of HRM and sequencing have moderate consistency, and HRM can also detect heterogeneous resistance.
Discussion
The detection of MTB drug sensitivity is essential for the control of TB, especially for the transmission of M/XDR MTB strains. Although the L-J proportion method is the gold standard method and is possible to detect 1% of drug resistant bacteria [14], it is difficult to meet the requirements of rapid detection because of slow growth of MTB, thus makes it not efficient for the treatment and transmission control, and the clinical conditions of TB patients maybe worsened during the long detective period. To popularize the early and sensitive diagnostic techniques is beneficial for the detection of strains with a low proportion of drug-resistant subgroup in the sample, so as to achieve the purpose of rapid detection of drug-resistant MTB. This study evaluated the performance of HRM as a rapid, accurate, simple and economical molecular biological technology for the detection of mutated genes in drug-resistant TB.
In this study, HRM was utilized for the detection of mutations in KatG315. A commonly mutation of KatG315 codon is AGC→ACC [15]. While a potential weakness of HRM is the detections of C→ G or T →A, which resulted from the Tm differences are usually less than 0.4ºC and 0.2ºC [16-20]. One possible improvement is to add another DNA for mixture, since the Tm of the heterologous chain will be lower than that of homozygous mutations, at which the peak curve is changed and is easier for discrimination [18,21- 23]. There are two ways for another (eg. H37Rv) DNA adding: before or after PCR amplification. If the DNA of H37Rv is added before PCR, the DNA concentrations of samples need to be controlled carefully, and all samples should be conducted the program twice: for adding and without adding H37Rv DNA. If DNA adding after PCR, only equal volumes of H37Rv and samples’ PCR products are required [24], and second HRM need to be performed for single peak samples. That is why we performed the method by adding H37Rv PCR products after first PCR for single curve samples. There were 11 strains were found mutations by HRM rather than sequencing. One possible reason was the emerging of heterogenous resistance, which means the concurrent presence of drug-sensitive and drugresistant MTB strains in one patient. At the beginning of treatment, the susceptible organisms were killed gradually, and the drug-resistant ones were increasing correspondingly. When reaching the threshold of detection [14], it was diagnosed as drug-resistant tuberculosis. Van Rie [25] found that these patients with mixed drug-sensitive and resistant MTB infections had poor response to treatment. At the same time, mixed infections can increase the chances of making disease course long and persistent, which may provide an opportunity to accumulate additional mutations and form stable, highlyresistant strains [26]. So, heterogeneous drug resistance could be considered as the initial stage of complete drug resistance [27]. Meanwhile, the discovery of heterogeneous drug resistance is beneficial for effective treatment timely by adjusting therapeutic protocols. Therefore, improving the detective ability of MTB heterozygous resistance has important clinical significance [28]. The conventional sequencing has low sensitivity for detecting heterogeneous resistance as more than 20% mutations demanded [29]. Study has revealed that the sensitivity of HRM is higher than DNA sequencing. The COLD-PCR COLDPCR/ HRM can detect as less as 5% of mutations [30]. Therefore, when mutations observed by HRM but not sequencing, it may suggest the number of mutations is lower than the limit of sequencing needed [6].
HRM used in this study has several advantages. Firstly, the results of heterogeneous resistant isolates can be given preferentially. Secondly, the DNA samples of MTB can be extracted by boiling, which is easily available at grass-roots units. Thirdly, the changes of peak curves are more obvious than the differences of Tm value. Fourthly, no additional reagents are required when the second HRM is performed. However, HRM has its limitation, for example, if the strain has neutralizing mutations in detected region, the curve is consistent with H37Rv [18]. In addition, more attentions should be paid for avoiding cross-contamination.
To increase efficacy of HRM for detecting drug-resistant mutations, future work will be focused on: improving DNA quality and putting HRM into practice among other anti-tuberculosis drugs and clinical specimens directly. With the deeper understanding of drug resistance mechanism in the future, drug sensitivity analysis that based on gene detection will be improved and HRM will be more optimized.
Conclusion
The HRM analysis involved in this study is a time-saving, low-cost, easyto- judgment method, and it has a powerful ability to detect heterogenous resistant strains. It is a suitable approach to be popularized in the basic medical institutions in Qinghai province for the detection of INH-resistant gene mutations.
Funding
This work was supported by grant from the National Natural Science Foundation of China (No.81860593)
Acknowledgments
We thank the stuff of Qinghai CDC for providing the MTB isolates and conducting L-J drug-sensitive tests.
REFERENCES
- Global tuberculosis report. Geneva: World Health Organization 2017.
- Prasad R. Multidrug and extensively drug-resistant TB (M/XDR-TB): problems and solutions. J Indian J Tuberc. 2010;57(4):180-91.
- Luo T, Jiang L, Sun W, et al. Multiplex real-time PCR melting curve assay to detect drug-resistant mutations of Mycobacterium tuberculosis. J Clin Microbiol. 2011;49(9):3132-38.
- Ong DC, Yam WC, Siu GK, et al. Rapid detection of rifampicin- and isoniazid-resistant Mycobacterium tuberculosis by high-resolution melting analysis. J Clin Microbiol. 2010;48(4):1047-54.
- Yi H, Hoffner S, Jiang W, et al. Extensive transmission of isoniazid resistant M. tuberculosis and its association with increased multidrug-resistant TB in two rural counties of eastern China: A molecular epidemiological studyJ. Bmc Infectious Diseases. 2010;10(1):43.
- Vossen RH, Aten E, Roos A, et al. High-resolution melting analysis (HRMA): more than just sequence variant screening. Hum Mutat. 2009;30(6):860-66.
- Nagai Y, Iwade Y, Hayakawa E, et al. High resolution melting curve assay for rapid detection of drug-resistant Mycobacterium tuberculosis. J Infect Chemother. 2013;19(6):1116-25.
- Choi G E, Lee S M, Yi J, et al. High-resolution melting curve analysis for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis clinical isolates. J Clin Microbiol. 2010;48(11):3893-98.
- Campbell PJ, Morlock GP, Sikes RD, et al. Molecular detection of mutations associated with first- and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2011;55(5):2032-41.
- Haeili M, Fooladi AI, Bostanabad SZ, et al. Rapid screening of rpoB and katG mutations in Mycobacterium tuberculosis isolates by high-resolution melting curve analysis. J Indian J Med Microbiol. 2014;32(4):398-403.
- Liu Y, Jiang X, Li W, et al. The study on the association between Beijing genotype family and drug susceptibility phenotypes of Mycobacterium tuberculosis in Beijing. J Sci Rep. 2017;7(1).
- CanettiG, Froman S, Grosset J, et al. Mycobacteria: laboratory methods for testing drug sensitivity and resistancej. Bull World Health Organ. 1963;29:565-78.
- Osman F, Ismail F, Osman A, et al. High Resolution Melting Curve Analysis for Rapid Detection of Pyrazinamide Resistance in Mycobacterium tuberculosis Clinical Isolates. J Tuberculosis Research. 2016;04(4):155-72.
- Rinder H. Hetero-resistance: an under-recognised confounder in diagnosis and therapy? J Med Microbiol. 2001;50(12):1018-20.
- Ahmad S, Mokaddas E. Contribution of AGC to ACC and other mutations at codon 315 of the katG gene in isoniazid-resistant Mycobacterium tuberculosis isolates from the Middle East. Int J Antimicrob Agents. 2004;23(5):473-79.
- Ramirez MV, Cowart KC, Campbell PJ, et al. Rapid detection of multidrug-resistant Mycobacterium tuberculosis by use of real-time PCR and high-resolution melt analysis. J Clin Microbiol. 2010;48(11):4003-9.
- Chen X, Kong F, Wang Q, et al. Rapid Detection of Isoniazid, Rifampin, and Ofloxacin Resistance in Mycobacterium tuberculosis Clinical Isolates Using High-Resolution Melting Analysis. Journal of Clinical Microbiology. 2011;49(10):3450-57.
- Pietzka AT, Indra A, Stoger A, et al. Rapid identification of multidrug-resistant Mycobacterium tuberculosis isolates by rpoB gene scanning using high-resolution melting curve PCR analysisJ. J Antimicrob Chemother. 2009;63(6):1121-27.
- Malhotra B, Goyal S, Bhargava S, et al. Rapid detection of rifampicin resistance in Mycobacterium tuberculosis by high-resolution melting curve analysis. Lung Disease the Official Journal of the International Union Against Tuberculosis & Lung Disease. 2015;19(12):1536-41.
- Mayerhofer B, Stoger A, Pietzka AT, et al. Improved protocol for rapid identification of certain spa types using high resolution melting curve analysis. PLoS One. 2015;10(3):e116713.
- Gundry CN, Vandersteen JG, Reed GH, et al. Amplicon melting analysis with labeled primers: a closed-tube method for differentiating homozygotes and heterozygotes. J Clinical Chem. 2003;49(3):396-406.
- Bruzzone CM, Tawadros PS, Boardman LA, et al. Enhanced Primer Selection and Synthetic Amplicon Templates Optimize High-Resolution Melting Analysis of Single-Nucleotide Polymorphisms in a Large Population. Genetic Testing & Molecular Biomarkers. 2013;17(9):675-80.
- Ong DC, Yam WC, Siu GK, et al. Rapid detection of rifampicin- and isoniazid-resistant Mycobacterium tuberculosis by high-resolution melting analysis. J Clin Microbiol. 2010;48(4):1047-54.
- Liew M, Pryor R, Palais R, et al. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. J Clin Chem. 2004;50(7):1156-64.
- Van Rie A, Victor T C, Richardson M, et al. Reinfection and mixed infection cause changing Mycobacterium tuberculosis drug-resistance patterns. Am J Respir Crit Care Med. 2005;172(5):636-42.
- Sergeev R, Colijn C, Cohen T. Models to understand the population-level impact of mixed strain M. tuberculosis infections. J Theor Biol. 2011;280(1):88-100.
- Hofmann-Thiel S, Van Ingen J, Feldmann K, et al. Mechanisms of heteroresistance to isoniazid and rifampin of Mycobacterium tuberculosis in Tashkent, Uzbekistan. Eur Respir J. 2009;33(2):368-74.
- Folkvardsen DB, Thomsen VO, Rigouts L, et al. Rifampin heteroresistance in Mycobacterium tuberculosis cultures as detected by phenotypic and genotypic drug susceptibility test methodsJ. J Clin Microbiol. 2013;51(12):4220-22.
- Pholwat S, Stroup S, Foongladda S, et al. Digital PCR to detect and quantify heteroresistance in drug resistant Mycobacterium tuberculosis. PLoS One. 2013;8(2):e57238.
- Pang Y, Liu G, Wang Y, et al. Combining COLD-PCR and high-resolution melt analysis for rapid detection of low-level, rifampin-resistant mutations in Mycobacterium tuberculosis. J Microbiol Methods. 2013;93(1):32-36.