Nucleocytoplasmic trafficking
- *Corresponding Author:
- Dr Grant N Pierce
Institute of Cardiovascular Sciences, Albrechtsen Research Centre, St Boniface Hospital, 351 Tache Avenue, Winnipeg, Manitoba R2H 2A6.
Telephone: 204-235-3206
Fax: 204-235-0793
E-mail: gpierce@sbrc.ca
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact support@pulsus.com
[ft_below_content] =>Keywords
Nuclear pore complex; Nuclear transport
Nucleocytoplasmic Trafficking
Nuclear transport is a complex process involving soluble, cytosolic machinery and membrane-bound components. Molecules that localize from cytosolic to nucleoplasmic compartments and vice versa must cross both the outer and inner nuclear membranes of the nuclear envelope (NE) to reach their destination. For this purpose, a large multiproteinaceous structure known as the nuclear pore complex (NPC) spans both membranes of the NE and serves as the gateway through which molecules transit [1-10].
The NPC
Structure and composition
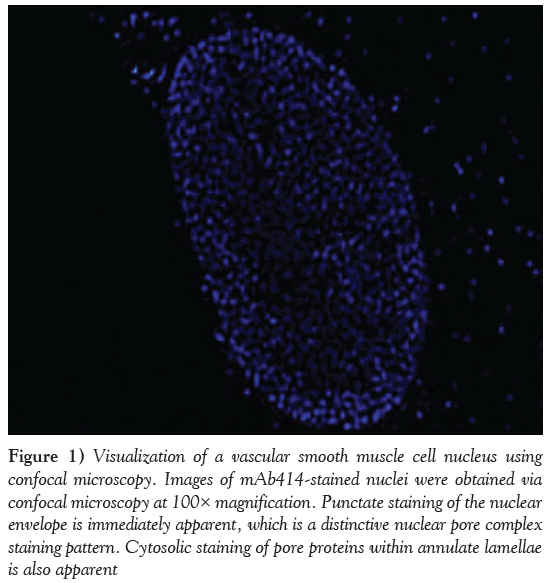
Electron microscopy of an NE in cross-section demonstrates how the NPC serves as a proteinaceous ‘rivet’ connecting both outer and inner nuclear membranes of the NE. Immunocytochemistry using mAb414, an antibody that recognizes NPC proteins or nucleoporins (nups), demonstrates intense staining of the nucleus as well as cytoplasmic annulate lamellae (Figure 1). Confocal microscopy of smooth muscle cells illustrates regular distribution of NPCs across the surface of the NE. The NPC is a dynamic and highly regulated structure, which shows a high degree of evolutionary conservation across various species [7]. Most of the information on NPC composition and architecture has been garnered from yeast [10] and Xenopus [11,12]; however, the primary focus of this section will be on mammalian NPCs.
Figure 1: Visualization of a vascular smooth muscle cell nucleus using confocal microscopy. Images of mAb414-stained nuclei were obtained via confocal microscopy at 100× magnification. Punctate staining of the nuclear envelope is immediately apparent, which is a distinctive nuclear pore complex staining pattern. Cytosolic staining of pore proteins within annulate lamellae is also apparent
Mammalian NPCs are approximately 125 MDa and composed of 30 to 32 discrete nups that occur in redundant groupings within the pore [7]. Overall, the NPC is approximately 120 nm in diameter and possesses an axial depth of approximately 70 nm. It is composed of eight multimeric subunits arranged in an annular configuration that can be visualized as a distinctive barrel-like shape. This structural conformation leaves a functionally dynamic aqueous central channel approximately 9 nm to 11 nm in diameter under normal conditions [13,14]. Electron microscopy of prepared NEs illustrates the octagonal symmetry of the pore when viewed en face. The central pore can dilate to accommodate larger cargoes in transit [15,16] as well as respond to calcium fluxes [17,18]. In addition to the main central channel [19,20], peripheral channels exist, which are hypothesized to permit the flow of various small molecules and ions [4,21]. One feature of the NPC core that remains controversial is the pore plug or central transporter. In some preparations, electron and atomic force microscopy have revealed the presence of a large molecule occluding the central channel of the pore [1]. It is unclear whether this plug is a bonafide structural component of the pore or simply trapped cargo; however, observations made by Stoffler et al [22] and others [23,24] suggest that the central plug may be a composite of a molecule caught in transit with attendant changes in distal ring conformation.
On the nuclear and cytoplasmic faces of the pore are ring structures from which filaments extend into their respective compartments [4]. The cytoplasmic and nuclear ring-filament assemblies are asymmetrical and possess distinct structural variations. The cytoplasmic filaments are regularly spaced around the ring and extend into the cytosol for approximately 50 nm [22]. They also contain nups, which serve as docking sites for pretransport import complexes [25-28].
The nuclear filaments are evenly distributed around the nuclear ring, which in turn interacts with the nuclear lamina [29]. The nuclear filaments extend significantly further into the nucleoplasm than their cytoplasmic counterparts and are joined at their distal ends by a smaller ring, termed the distal ring [3,4,22,29]. This entire assembly is known as the ‘nuclear basket’ [3,4]. Sites on the distal ring and at the distal ends of the nuclear filaments serve as initial docking sites for export complexes and serve a function similar to their cytosolic counterparts [30]. Another unique feature of the nuclear face of the NPC is its association with the nuclear lamina (via the nuclear ring) [29]. The function of the nuclear lamina in this respect is to anchor the NPCs within the NE [31].
Dynamics
The nuclear pore is a dynamic structure that responds to a variety of stimuli [32-35], and its functional diversity extends beyond its role as a transport gate, discussed in excellent reviews elsewhere [36,37]. The presence or absence of the central transporter in response to the depletion or repletion of periplasmic calcium stores, respectively, is an excellent example of NPC dynamics [17,18]. It has been proposed that this transporter – or ‘plug’ – may represent a calcium-sensitive multimer composed of nup gp210 [32]. Others have suggested that this feature is a preparative artifact because it was not observed in other experiments and was postulated to be molecular cargo trapped in transit (35). Numerous studies investigating the role of calcium in NPC regulation, however, have demonstrated a calcium-sensitive mechanism responsible for closing and opening the pore [17,18]. These independent observations support the notion that the NPC is dynamically regulated by calcium fluxes in and around the nucleus [17,18,38].
Nups are capable of diffusing from one membrane system to another within the same cell. Partial pore complexes and individual nups have been localized to the cytoplasmic annulate lamellae (AL), an organelle associated with components of the Ran cycle and potentially involved in nup distribution [39-41]. Morphologically, the AL are multilayered membrane structures [31,41,42]. Harel et al [40] demonstrated that imbalances in cellular RanGTP concentrations lead to excessive formation of de novo AL; therefore, the presence of AL may be a secondary effect of nuclear assembly processes. However, the identification of partial pore complexes within the AL suggests that the AL may serve as a distributor and repository for pre-assembly NPC proteins and nuclear pore subcomplexes [41]. This was based on the observation that immunofluorescent staining for nups visualized proteins within the NPC as well as within the AL [43]. Belgareh and Doye [44] reported transnuclear diffusion of a nuclear pore protein in a binucleate heterokaryon, demonstrating that some nups are capable of translocating from one membrane system to another. This finding provides evidence of the hypothetical role of the AL as a nup repository and supports the dynamic characteristic of the NPC. Interphasic NPCs have low turnover in living cells and are replaced after a round of mitosis [31]. Significantly, individual nups have variable half-lives [45], with some nups demonstrating off-pore mobility [31,46]. In untreated cells, individual NPCs possess limited lateral movement and show heterogeneous distribution because they are anchored to the nuclear lamina via the nuclear ring [31]. They are capable, however, of shifting in waves around the entire surface of the nucleus as a single entity [31], which can be envisioned as a net covering a ball (ie, although the entire net can freely move around the surface of the ball, the interconnecting points are not significantly displaced with respect to one another). Abnormal spatial distribution of NPCs has been reported in conjunction with alterations or defects in nuclear lamins [47,48]. Mutations in a specific subset of nups can also result in NPC clustering instead of the normal distribution [44,49-51]. These observations imply that, although NPCs maintain specific distributions under normal conditions, they are capable of collectively undergoing significant spatial rearrangement and are not static structures. Additionally, the density of NPCs can change according to the metabolic needs of the cell [52-54]. Changes in the nuclear pore can, thus, be correlated with growth requirements and/or changes in mitosis.
The link between NPC dynamics and mitosis has been further strengthened by work that has demonstrated the redistribution of integral nups to the kinetochore during mitosis [55-57]. The trigger responsible for relocalizing these NPC proteins has been presumed to be NE breakdown [57], which releases the nups, enabling them to associate with the mitotic machinery [57]. The cell can then proceed past the spindle checkpoint and subsequent steps in mitosis can continue unimpeded. From these observations, it is clear that individual nuclear pore proteins are mobile in both interphase and mitosis, significantly more so in the latter.
Models of Transport
Despite our present knowledge of the structure and characteristics of the nuclear pore, the nature of the barrier that regulates passage of molecules across the NPC remains to be clearly defined. Early research in yeast demonstrated that some nups contained FG repeats as well as GLFG and FxFG motifs [58]. These sequences are highly conserved between NPCs of different species and facilitate searches for homologous proteins in higher eukaryotic species. Importantly, the interaction of nups with soluble components of the nuclear transport machinery occurs through these FG repeats [59-61] and forms the basis of several models of nuclear transport.
Polymer brush
A thorough analysis of the yeast NPC by Rout et al [10] revealed that the yeast NPC contains approximately 30 to 32 nups. Based on the number and distribution of nups observed in their work, Rout et al [10] suggested Brownian affinity as a potential mechanism of transport. The notion of transport here was that import or export complexes bind to FG nups within the NPC, increasing their residence time at the pore opening, thus raising the probability of their entry into the channel and subsequent translocation through the central channel by Brownian motion [10]. This initial understanding served as the conceptual precursor to the present polymer brush model currently proposed
In this model, disordered FG domains extend to create a virtual gate, or entropic brush region that blocks non-NLS-mediated protein movement [62]. In contrast, binding via importin-b collapsed the extended domain. Furthermore, this effect was reversed by the action of RanGTP that would return the collapsed regions to their extended, brush conformation [63].
hydrogel
Another model that attempts to take into account cargo movement is the hydrogel model [64,65]. This model hypothesizes the existence of an interconnected meshwork between nups that are connected to one another via their FG repeats within the channel [64]. Transport cargoes are able to traverse this meshwork because they are able to interact with the FG repeats themselves, thus breaking the connection between nups within the mesh. Additionally, the ‘holes’ within the mesh act as a molecular sieve and together with the hydrophobic nature of the channel serve to exclude both inert (receptor-less) cargo and molecules beyond the size limit of the pore [64].
Forest (trees and bushes) model
A third proposed mechanism of action focuses on the bimodal distribution of two categories of extended FG conformations and their arrangement within the NPC (66). In this model, FG nups with extended disordered domains (trees) or relatively compact FG regions (bushes) are arranged in a manner that creates distinct transport zones within the central channel. This combines aspects of the polymer brush and hydrogel models, and takes into account the varying affinities of discrete nups for specific cargo-receptor complexes.
The three models described above explain the movement of cargoes across the NE; however, the role of the Ran gradient is not fully considered for each. The polymeric brush model does provide evidence for the role of RanGTP in altering permeability, but it is a fourth model that attempts to explain the selective permeability barrier of the NPC in the context of a full Ran gradient. This is critical to nuclear transport processes, as collapse of the gradient can stop transport [67-70]. Additionally, the direction of transport can be reversed in the presence of high cytosolic concentrations of RanGTP [71].
Karyopherin-centric, binding affinity model
Ben-Efraim and Gerace proposed a model that would incorporate the directionality and motive force provided by RanGTP/GDP distribution across the NE with size exclusion limits and binding affinities [72]. In their model, transport cargoes bound to RanGTP move up an increasing affinity gradient to end up at terminal docking sites in their predestined compartment [72-76]. This model reconciled biophysical parameters of the NPC with the observed transport dynamics, and would ultimately serve as the conceptual precursor of the contemporary karyopherin-centric model [77,78]. This model incorporates the observation that specific FG-containing nups bind transport receptors with varying degrees of affinity [72].
While all proposed models explain the exclusivity of the selective FG nup gating mechanism to some degree, no consensus has yet been reached that unifies the spectrum of biophysical properties exhibited by the NPC barrier with the diversity of biochemical factors critical for nucleocytoplasmic transport. Future work using technology capable of increased NPC structural detail and high temporospatial resolution [79,80] of discrete transport events will contribute greatly to addressing this long-standing challenge within the field.
Nuclear Transport Signal Sequences
Nuclear localization signals
Nuclear transport generally describes the process by which molecules move back and forth between the nuclear and cytoplasmic spaces. It is a bidirectional and energy-dependent phenomenon, with specific regulatory mechanisms distributed within the soluble portion of the cell as well as at the NE. Nucleocytoplasmic trafficking is initiated on recognition of a specific signal sequence that directs the molecule to either be imported into the nucleus from the cytoplasm or exported in the opposite direction.
The first signal sequence identified was a monopartite, polybasic amino acid motif (PKKKRKV) directly responsible for the nuclear localization of the SV40 large T antigen [81]. This nuclear localization signal (NLS) was a polylysine motif known as the ‘classical’ signal sequence (Table 1). NLSs also exist in a bipartite form. Dingwall et al [82] first characterized the existence of a bipartite NLS in nucleoplasmin and would later demonstrate that it contained polybasic sequences homologous to the NLS of the SV40 large T antigen [83].
| Import receptor | Signal sequence/motif |
|---|---|
| Importin-α | (monopartite) PKKKRKV (3) |
| (bipartite) KRPAAIKKAGQAKKKK (7) | |
| Importin-β | RQARRNRRRRWR (8) |
| Transportin | NQSSNFGPMKGGNFGGRSSGPY |
| GGGGQYFAKPRNQGGY (8) | |
| Snurportin1 | trimethylguanosine cap (m3G) (9) |
| XRIPα | DNA binding domain (10) |
Importin-α is the primary receptor for the ‘classical’ polybasic amino acid motif, which occurs as a monopartite or bipartite signal sequence (see text and above). Once importin-α binds its substrate, an importin-β binding domain on the alpha subunit is exposed, leading to the formation of a heterotrimeric receptor-substrate complex. Importin-β is also a primary receptor and can bind to individual proteins without the need of an adaptor protein. Other import receptors recognize nuclear localization signals distinct from the polylysine type and are depicted above
Table 1: Examples of nuclear import sequences and their cognate receptor
The discovery of the NLS provided the impetus for the identification of other signal sequences responsible for nuclear import, but distinct from those of the SV40 variety. A glycine rich, novel sequence that did not contain any ‘classical’ NLS-type basic amino acids was found in the pre-messenger/mRNA binding protein hnRNP A1 [84] and was designated M9 and recognized by transportin (Table 1). Attachment of this sequence to two cytosolic enzymes, pyruvate kinase and beta-galactosidase, was sufficient to enable their import into the nucleus. A listing of selected NLSs and their cognate receptors are shown in Table 1.
Nuclear export signals
Proteins and RNA exported from the nucleus typically possess a leucinerich signaling motif known as the nuclear export signal (NES). Similar to NLSs, a variety of signaling motifs exist that are capable of mediating nuclear export [85]. Instead of amino acid sequences, RNA species possess structural motifs recognized by specialized exportin molecules that direct them toward the cytoplasm [86]. Examples of various NES are shown (Table 2).
| export receptor | Signal sequence/motif | Molecule exported |
|---|---|---|
| Crm1 | LPPLERLTLD (20) | Rev |
| LALKLAGLDI (21) | PKI | |
| LQKKLEELELE (22) | MEK1 | |
| CAS | CGGLDKIE (25) | importin-α |
| Calreticulin | CGGGKVFFKRAVEGQHNLY (26) | glucocorticoid receptor |
| TAP | RNA stem-loop structure (23) | RNA |
| Mex67p | Polyadenylated RNA (28) | |
Exportins are responsible for delivering proteins to the cytosol from the nucleus. Crm1 (also known as exportin-1) has been well characterized as a nuclear exporter. Other proteins have been identified that bind signals distinctly different from those recognized by Crm1. RNA molecules are exported from the nucleus once their receptor recognizes distinct molecular features (see above)
Table 2: Selected nuclear export signals and their receptors
Due to the variety of transport signals available for import and export, nucleocytoplasmic trafficking remains a tightly controlled process. The primary level of regulation occurs at the nuclear transport signal by controlling access to the NLS/NES. With respect to the localization signal, the eukaryotic cell has evolved various mechanisms to control nuclear trafficking. Phosphorylation, dephosphorylation and regulated proteolysis are some of the common regulatory processes that play a role in regulating nuclear transport at one or more levels [87-92].
Regulation of NlSs and NeSs
A common method of regulation within the eukaryotic cell is phosphorylation of a target substrate. In all phosphorylation and dephosphorylation cycles, there are two classes of enzymes responsible for adding and removing phosphate groups, called kinases and phosphatases, respectively.
A diverse number of kinases exist that can be regulated by cyclic AMP and GMP [93-95], calcium [96] and lipids [97-99]. The process of phosphorylation begins when a protein substrate possesses a motif recognized by the kinase. On association of the kinase with its target, ATP is recruited to the complex and the terminal phosphate (designated as the γ phosphate) is transferred to the recipient protein, generating the phosphoprotein, ADP or AMP, plus an inorganic phosphate as reaction products. The amino acids phosphorylated in mammalian systems are referred to as O-phosphates and include tyrosine, threonine and serine [100]. O-phosphates are phosphoproteins in which the hydrogen on the hydroxyl moiety of the R-group has been substituted with an anionic phosphate group. More than one of these phosphoaminoacids within a given protein may be phosphorylated, as is the case with activated MAP kinases, which are dually phosphorylated on both threonine and tyrosine residues [101]. While the three O-phosphates are, by far, the most commonly studied, other phosphoaminoacids, such as phosphohistidine, also exist [100]. Phosphoaminoacids that flank the nuclear localization signal within a protein can affect the affinity of a transport receptor for the NLS on phosphorylation. For example, phosphorylation of the residues adjacent to the NLS of the SV40 large T antigen regulates nuclear import of the large T antigen [102].
The removal of phosphate groups is performed by protein phosphatases [103]. Phosphatases can be regulated by calcium and calmodulin [104] and can be classified into four different families based on substrate specificity and degree of conservation within their catalytic domains [105]. With respect to nuclear transport, dephosphorylation functions in a manner similar to that of phosphorylation. In this case, the removal of a phosphate group can unmask the transport motif, leading to its recognition and binding by the appropriate transport receptor. For example, dephosphorylation of NFAT exposes its NLS, subsequently leading to its nuclear translocation [87]. Furthermore, recent work has identified an association between protein phosphatase 2A and members of the importin-β superfamily [106], which may indicate the existence of generalized phosphatase-dependent regulation on nuclear transport machinery.
Regulation of nucleocytoplasmic transport occurs within the cytosol and possibly at the NE. Both kinases and phosphatases have been identified to reside within the nuclear pore [107]. A brief overview of transport receptors, their subclasses, and a short review on the NPC are presented in the following sections.
Nuclear Transport Receptors
The proteins responsible for the movement of molecules back and forth between the nuclear and cytosolic compartments are collectively referred to as importins [108]. Identification of the first importins, also known as karyopherins, occurred in the early 1990s. It was the isolation and characterization of importin-β (or karyopherin β1) [109] that unequivocally demonstrated the requirement of a cytosolic factor in the nuclear import process. As more transport receptors were discovered in subsequent years, it became apparent that all of them could be categorized into one family. The importin-β superfamily is the de facto designation describing these related proteins. For mechanistic reasons, transport receptors responsible for the export of molecules out of the nucleus into the cytoplasm also fall within this importin superfamily [108].
Importin-β
The prototypical importin-β protein was first isolated and identified as a 97kDa molecule in bovine erythrocytes [109], and was subsequently found to be essential to nuclear protein import [109,110]. Structural analyses of importin-β revealed that it contains 19 tandem HEAT repeats, arranged so that the tertiary conformation of the molecule is a right-handed superhelical protein with a high degree of flexibility [111]. Functionally, it can bind to nuclear pore proteins containing the FxFG motif and possesses an amino-terminal Ran binding domain [112-114]. With respect to nuclear import, importin-β participates as part of a trimeric complex formed when a protein bearing a ‘classical’ polybasic NLS is recognized by its receptor, importin-α [115]. Interaction of the NLS with its cognate receptor causes importin-α to undergo a conformational shift which then promotes its binding to the carboxy terminus of importin-β by virtue of an importin-β binding (IBB) domain at its N-terminus [111,116]. Formation of the importin heterodimer causes mutual conformational changes in both subunits which ultimately promote nuclear protein import [116] in a Ran and energy dependent manner.
Alternatively, a protein may directly bind importin-β without the use or need of adaptor proteins such as importin-α [108]. Several viral proteins [117-119] are able to bind directly to the IBB domain and form a heterodimeric complex which is imported into the nucleus. Additionally, other endogenous eukaryotic proteins use this mechanism to be imported [120,121]. Based on these observations, the importin-β superfamily can be subdivided into two categories: those that are capable of binding a molecule directly for import/export; and those that utilize adaptor proteins. Adaptor proteins themselves can be further classified into importin-α type NLS receptors, or nonimportin- α type receptors.
Adaptors: importin-α
Importin-α is the prototypical adaptor protein that recognizes both mono- and bipartite ‘classical’ NLSs [122,123] and was simultaneously isolated and identified with importin-β, as mentioned previously (109). This molecule consists of 10 tandem ARM motifs, with the main NLS recognition sites residing within the second and fourth ARM repeats [124]. At the C-terminus is an acidic domain required for binding to CAS, the endogenous nuclear exporter of importin-α [125-127]. At its N-terminus, importin-α possesses the IBB domain required for interaction with importin-β [111,127].
Identification of a polybasic sequence within the IBB [128] led to the hypothesis that importin-α may also possess autoinhibitory activity in addition to binding to importin-β. This was confirmed by a later study which reported that a Lys-Arg-Arg sequence within the IBB domain binds to the NLS recognition domain within importin-α [129]. This highly conserved sequence was identified in several forms of importin-α from yeast, mouse and human sources [129]. Using a variety of plasmids encoding different mutations of this IBB domain, Harreman et al [129] demonstrated that this sequence was essential in promoting NLS-cargo release within the nucleus and it was suggested that the autoinhibitory properties of the IBB domain prevents futile cycling of unloaded importin-α/β complexes.
More than one form of importin-α exists. In humans, six forms have been identified, each of which are separate gene products [130], as is the case for the various mouse isoforms of importin-α that have been identified [131]. Since tissue specific distribution of the importin-α isoforms has been observed, it is possible that each isoform is responsible for the import of a specific group or subset of proteins. For example, RanGEF/RCC1 is a constitutively nuclear enzyme that is preferentially imported by importin-α3 [130,132]. Future studies investigating specific importins and their respective cargoes will provide a more comprehensive understanding of the diversity of import receptors involved in nuclear transport.
Adaptors: non-importin-α receptors
There are other adaptor proteins with specific cargoes distinct from those mediated by the importin α/β complex. In some ways, these molecules possess similar characteristics to those of importin-α. For example, they possess an IBB domain as well as the ability to bind to their respective cargoes. However, the recognition motif by which they bind is typically quite different.
Snurportin1 was identified as the adaptor responsible for importing m3G capped U snRNPs into nuclei [133]. Similar to importin-α, snurportin1 possesses an IBB domain. However, it is distinct from the former in that it allows Ran- and energy- independent nuclear protein import [134]. In order to illustrate that this ability was characteristic of the snurportin1 IBB, Huber et al [133] generated a snurportin1 mutant that possessed the importin-α IBB and demonstrated that ribonucleoprotein import became Ran dependent.
Histone H1 is a small (~22 kDa) basic protein that is consitutively nuclear and actively transported despite its small size [135-137]. Also known as linker histones, they are involved in both the maintenance of chromatin structure as well as transcriptional regulation [137] and are the exclusive ligands of the importin-7 receptor [136,138]. Several portions of histone H1 possess stretches of basic amino acids that, on their own, serve as NLSs that can functionally interact with individual import factors (136). However, import of the entire complex requires a heterodimeric receptor composed of both importin-β and importin-7, the latter being the adaptor specific for histone H1 [136]. It is believed that in addition to serving as an import receptor, the importin-β/-7 complex may also serve as a chaperone for histone H1 [136].
The XRIPα adaptor protein is responsible for importing replication protein A (RPA) [139]. RPA is a single stranded DNA binding protein composed of three subunits (140) that is essential for chromosomal DNA replication [141], repair [142] and recombination [143-145]. The adaptor was originally identified using a yeast two hybrid assay by Jullien et al [139]. Briefly, several domains of the RPA holoenzyme were fused to the Gal4 DNA-binding domain to form the “bait” constructs that were used to screen a Xenopus oocyte cDNA library. The identified adaptor was found to specifically bind RPA and possessed an IBB structurally distinct from that of the importin-α IBB [139]. Similar to previously characterized importin-β binding domains, the IBB of XRIPα is located at the N-terminus and contains an arginine-rich, basic motif [139]. The similarities end there, however, as arginines in this particular receptor do not form continuous stretches of basic amino acids as found in the IBBs of HIV Tat and Rev proteins [119].
It is clear from these examples that although members within the importin family share some properties, they possess individualized characteristics which contribute to the recognition of a broad and diverse array of substrates. While this observation has been made for molecules that mediate nuclear import, it also holds true for the members of the importin-β superfamily that are responsible for nuclear export.
Exportins
Exportins are members of the importin-β family that are responsible for the movement of proteins, RNA and ribonucleoproteins from the nucleus into the cytoplasm [107,146]. They are generally similar to the importins in that they recognize a localization signal, bind to Ran and interact with proteins of the NPC. On closer investigation of the exportin molecule, differences become apparent.
CRM1/exportin-1 is the prototypical exportin molecule responsible for nuclear transport of the bulk of exported substrates. It recognizes a nuclear export signal (NES), which is different from the polybasic NLS mentioned earlier [147]. NESs are typically leucine rich sequences found on a protein destined for transport into the cytosol [148]. The first such NES was identified in the HIV Rev protein (149). In nuclear export, CRM1 binds to the NES and associates with RanGTP to form a stable export complex, similar to the mechanism of nuclear import. This heterotrimeric complex is then directed to the nuclear basket on the nucleoplasmic face of the NPC [76] and subsequently exported to the cytoplasm. Once there, RanGAP activates the intrinsic GTPase activity of Ran [150]. Hydrolysis of GTP to form RanGDP promotes dissociation of the nuclear export complex, releasing the cargo into the cytoplasm. Although this pathway is recognized as the ‘workhorse’ pathway of nuclear export, other CRM1- independent transport mechanisms exist.
One such example is the nuclear export of importin-α. Kutay et al [126] identified CAS as an essential protein required to transport importin-α from the nucleus back to the cytosol following a round of nuclear import. Subsequent studies confirmed the functionality of CAS as a nuclear exporter [151-153]. CAS activity can be modulated by sphingolipids like ceramide to alter nuclear protein import [154]. Calreticulin is another nuclear export receptor identified by Holaska et al [155]. Although indirectly involved in nuclear import of NFAT3 [156] and MEF2C [157], calreticulin directly exports nuclear, steroid, non-steroid hormone and orphan receptors [158,159] from the nucleus in a calcium-dependent manner [160]. This observation was surprising, given the constitutive localization of calreticulin to the endoplasmic/sarcoplasmic reticulum [161]. Calreticulin possesses an endoplasmic reticulum (ER)/sarcoplasmic reticulum retention signal which limits its presence in the cytosol [161]. Walther et al [158] examined calreticulin-mediated export of the glucocorticoid receptor (GR) using a heterokaryon assay and reported that transient permeabilization of the ER membrane allowed calreticulin to leave the ER lumen. Calreticulin release led to increased nuclear export of GR compared with rates observed in control cells and it was hypothesized that calreticulin-mediated export was a normal physiological mechanism used to rapidly clear the nucleus of receptors before mitosis [158]. The role of calreticulin as a nuclear export receptor remains debatable because no identifiable nuclear localization signal or import mechanism has been characterized for calreticulin.
Proteins are not the only molecules subject to exportin mediated transport. The export of RNA species involves completely different sets of receptors. The transport of transfer RNAs (tRNAs) is mediated specifically by exportin-t [162,163] and to a lesser extent, exportin-5 [164]. Both exportin-t and exportin-5 cooperatively bind RanGTP and the tRNA molecule directly, without the need for adaptors. Similar to tRNAs, mRNAs possess specific proteins which direct them out of the nucleus in a CRM1- and Ran- independent manner.
Messenger RNA is delivered to the cytosol by the TAP receptor [165]. This exportin forms a complex with another molecule, NXT1, to form a heterodimer that mediates the movement of mRNA out of the nucleus [166-168]. However, transport of mRNA is slightly more complex, as other factors which participate in splicing, elongation and termination of the nascent mRNA transcript are required to mediate the association between the TAP/NXT1 complex and mRNA [146,169,170].
The diverse nature of nuclear transport receptors and adaptors justifies the premise of the existence of other receptors that remain to be identified [171]. Also, while knowledge concerning nucleocytoplasmic trafficking control has come primarily from studies that have examined regulatory processes on the cytosolic machinery, evidence suggests that modifications at the membrane bound NPC also regulate nuclear trafficking [172].
Ran and Nuclear Transport
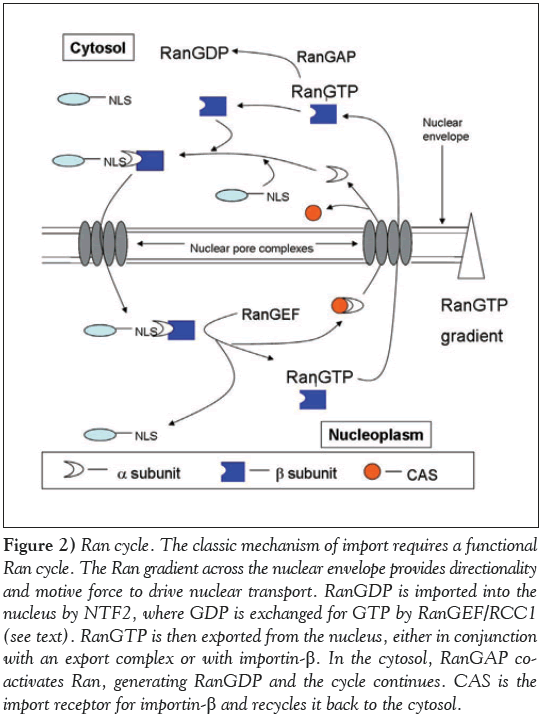
Ran is a small GTPase intimately involved with nucleocytoplasmic trafficking (Figure 2) [70,142,173,174]. Located predominantly in the nucleus, it is capable of shuttling back and forth between the nuclear and cytoplasmic compartments [175,176]. Nuclear Ran is bound to GTP and delivered to the cytosol in conjunction with an export complex [177]. Once there, it is converted to RanGDP [150,178]. This is extremely important, as hydrolysis of RanGTP promotes the dissociation of exportins from their cargo when they reach the cytosol.
Figure 2: Ran cycle. The classic mechanism of import requires a functional Ran cycle. The Ran gradient across the nuclear envelope provides directionality and motive force to drive nuclear transport. RanGDP is imported into the nucleus by NTF2, where GDP is exchanged for GTP by RanGEF/RCC1 (see text). RanGTP is then exported from the nucleus, either in conjunction with an export complex or with importin-β. In the cytosol, RanGAP coactivates Ran, generating RanGDP and the cycle continues. CAS is the import receptor for importin-β and recycles it back to the cytosol.
While cytoplasmic delivery of Ran occurs via its participation in an export complex, import of Ran requires the RanGDP specific import factor called p10/NTF2. First identified in S. cerevisiae by Nehrbass and Blobel [179], this molecule was shown by these and other authors to bind to the FxFG repeats of isolated nups (180,181). Further work clarified the role of p10/NTF2 as a nuclear import receptor for Ran in its GDP bound form [182-184], as it was observed that NTF2 specifically bound RanGDP and not RanGTP [179]. NTF2 is located primarily at the NE, bound to nups [181], since the Ran-binding and nup binding domains are discrete regions of the protein (180). It is also distributed between the cytoplasm and the nucleus [180]. The concentration of NTF2 at the NE is on the order of ~20 μM and it exists in a dimerized form, whereas it is monomeric in the cytoplasm and the nucleus, at a concentration of 0.3 μM and 0.6 μM, respectively [180]. These differences in concentration affect the monomer-dimer equilibrium of NTF2, as Chaillan- Huntington et al reported a dissociation constant of ~1.1 μM for NTF2 [180]. At concentrations below this KD, the dimer dissociates and significantly weakens the affinity of NTF2 for RanGDP [180], releasing it into the nucleoplasm, which can then be recharged with GTP by RCC1.
Ran has weak inherent GDP/GTP exchange activity as well as intrinisic GTP hydrolyzing capabilities [185]. In order for Ran to maintain its distinct GTP and GDP bound states in the nucleus and cytoplasm, respectively, a host of accessory proteins exist which regulate and enhance both the hydrolyzing ability of Ran as well as its nucleoside binding capacity. Within the nucleus is the RanGEF/RCC1 enzyme. It is constitutively imported and maintains its nuclear localization by associating with chromatin (186). This enzyme is responsible for replacing GDP with GTP, and maintains Ran in a GTP bound form [187]. Binding of RanGTP to importin-β entering the nucleus promotes dissociation of the import complex and subsequent release of the NLS bearing cargo into the nucleus [188]. Export complexes are formed only in the presence of RanGTP [177,189]. RCC1 is imported into the nucleus by two distinct mechanisms – one which depends on importin-α3 [130] and another that does not depend on energy or preexisting Ran gradients [190].
RanGTP that exits the nucleus in association with an export complex is converted to RanGDP by the concerted action of several coactivating proteins. Chief among these proteins is RanGAP1 [150], which, together with the Ran binding protein RanBP1 [191], increases the intrinsic hydrolytic activity of Ran. RanGAP1 exists in two forms, a soluble cytosolic form and a SUMOylated, NPCassociated form [91,92]. At the NPC, another Ran binding protein called RanBP2/Nup358 is part of the cytoplasmic filaments of the complex which also acts co-operatively with SUMO-RanGAP1 [91,92]. These elements maintain Ran in a GDP bound form in the cytoplasm. The nuclear components of the Ran cycle maintain high nuclear concentrations of RanGTP. It has been estimated that the concentration of RanGTP is over 200-fold greater in the nucleus than in the cytoplasm [192]. Together, the cytoplasmic and nuclear elements of the Ran cycle establish a steep RanGTP gradient across the NE. This is the source of energy for active nuclear transport as well as the element that specifies the vectorial movement of transport cargo.
The majority of nucleocytoplasmic trafficking requires a functional Ran cycle to operate (Figure 2). Exceptions exist, however, and Ran independent transport has been identified for a variety of different proteins [193,194]. The nuclear import receptor transportin enters the nucleus by a Ran unassisted mechanism [194]. IκBα, a regulator of NFκB transport, is imported without Ran [195]. Other non-transport proteins, such as beta catenin [196] and the U1A and U2B'' spliceosome proteins [197] also enter the nucleus by a pathway independent of Ran. RanGAP activity can be stimulated by lipid molecules like lysophosphatidylcholine [198].
The Future of Research on Nucleocytoplasmic Trafficking
Transport of proteins in and out of the nucleus is rapidly becoming recognized as a key process in the life and death of a cell [199]. It is also a critical pathway in the capacity of a cell to adapt to an everchanging environment [199]. With such a central role in the viability and function of a cell, it is not surprising that scientists are becoming increasingly aware of its potential role in cell pathology [200-203] and its identifications as a target for therapeutic interactions [204]. It is predicted that nucleocytoplasmic trafficking will become an increasingly important and attractive subject in cell biology.
Disclosures
The authors have no financial disclosures or conflicts of interest to declare.
References
- Pante N, Aebi U. Molecular dissection of the nuclear pore complex. Crit Rev Biochem Mol Biol 1996;31:153-99.
- Forbes DJ. Structure and function of the nuclear pore complex. Annu Rev Cell Biol 1992;8:495-527.
- Hinshaw JE. Architecture of the nuclear pore complex and its involvement in nucleocytoplasmic transport. Biochem Pharmacol 1994;47:15-20.
- Hinshaw JE, Carragher BO, Milligan RA. Architecture and design of the nuclear pore complex. Cell 1992;69:1133-41.
- Panté N, Aebi U. The nuclear pore complex. J Cell Biol 1993;122:977-84.
- Panté N, Aebi U. Exploring nuclear pore complex structure and function in molecular detail. J Cell Sci 1995;19(Suppl):1-11.
- Adam SA. The nuclear pore complex. Genome Biol 2001;2:1-6.
- Miller M, Park MK, Hanover JA. Nuclear pore complex: Structure, function, and regulation. Physiol Rev 1991;71:909-49.
- Ryan KJ, Wente SR. The nuclear pore complex: A protein machine bridging the nucleus and cytoplasm. Curr Opin Cell Biol 2000;12:361-71.
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J Cell Biol 2000;148:635-51.
- Panté N, Thomas F, Aebi U, Burke B, Bastos R. Recombinant Nup153 incorporates in vivo into Xenopus oocyte nuclear pore complexes. J Struct Biol 2000;129:306-12.
- Akey CW, Radermacher M. Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J Cell Biol 1993;122:1-19.
- Danker T, Oberleithner H. Nuclear pore function viewed with atomic force microscopy. Pflugers Arch 2000;439:671-81.
- Panté N, Kann M. Nuclear pore complex is able to transport macromolecules with diameters of ~39 nm. Mol Biol Cell 2002;13:425-34.
- Feldherr C, Akin D, Moore MS. The nuclear import factor p10 regulates the functional size of the nuclear pore complex during oogenesis. J Cell Sci 1998;111:1889-96.
- Feldherr C , Akin D, Cohen RJ. Regulation of functional nuclear pore size in fibroblasts. J Cell Sci 2001;114:4621-7.
- Perez-Terzic C, Jaconi M, Clapham DE. Nuclear calcium and the regulation of the nuclear pore complex. Bioessays 1997;19:787-92.
- Perez-Terzic C, Pyle J, Jaconi M, Stehno-Bittel L, Clapham DE. Conformational states of the nuclear pore complex induced by depletion of nuclear Ca2+stores. Science 1996;273:1875-7.
- Mazzanti M, Bustamante JO, Oberleithner H. Electrical dimension of the nuclear envelope. Physiol Rev 2001;1:1-19.
- Keminer O, Peters R. Permeability of single nuclear pores. Biophys J 1999;77:217-28.
- Shahin V, Danker T, Enss K, Ossig R, Oberleithner H. Evidence for Ca2+- and ATP-sensitive peripheral channels in nuclear pore complexes. FASEB J 2001;15:1895-901.
- Stoffler D, Feja B, Fahrenkrog B, Walz J, Typke D, Aebi U. Cryo-electron tomography provides novel insights into nuclear pore architecture: Implications for nucleocytoplasmic transport. J Mol Biol 2003;328:119-30.
- Sharma A, Solmaz S R, Blobel G, Melcak I. Ordered regions of nucleoporins Nup62, Nup54, and Nup58 form dynamic complexes in solution. J. Biol Chem 2015; 290:18370-8.
- Solmaz S R, Blobel G, Melcak I. Ring cycle for dilating and constricting the nuclear pore. Proc Natl Acad Sci USA 2013;110:5858-63.
- Yokoyama N, Hayashi N, Seki T, et al. A giant nucleopore protein that binds Ran/TC4. Nature 1995;376:184-8.
- Yaseen N R, Blobel G. Two distinct classes of Ran-binding sites on the nucleoporin Nup-358. Proc Natl Acad Sci USA 1999;96:5516-21.
- Yaseen N R, Blobel G. GTP hydrolysis links initiation and termination of nuclear import on the nucleoporin Nup358. J Biol Chem 1999;274:26493-26502.
- Wu J, Matunis M J, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran- GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine rich region. J Biol Chem 1995;270:14209-13.
- Goldberg M W, Allen T D. High resolution scanning electron microscopy of the nuclear envelope: Demonstration of a new, regular, fibrous lattice attached to the baskets of the nucleoplasmic face of the nuclear pores. J Cell Biol 1992;119:1429-40.
- Fahrenkrog B, Maco B, Fager AM, et al. Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J Struct Biol 2002:140:254-67.
- Daigle N, Beaudouin J, Hartnell L, et al. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J Cell Biol 2001;154:71-84.
- Moore-Nichols D, Arnott A, Dunn RC. Regulation of nuclear pore complex conformation by IP3 receptor activation. Biophys J 2002;83:1421-8.
- Perez-Terzic C, Gacy A M, Bortolon R, et al. Structural plasticity of the cardiac nuclear pore complex in response to regulators of nuclear import. Circ Res 1999;84:1292-301.
- Goldberg MW, Rutherford SA, Hughes M, et al. Ran alters nuclear pore complex conformation. J Mol Biol 2000;300:519-29.
- Stoffler D, Goldie K N, Feja B, Aebi U. Calcium-mediated structural changes of native nuclear pore complexes monitored by time-lapse atomic force microscopy. J Mol Biol 1999;287:741-52.
- Pascual-Garcia P, Capelson M. Nuclear pores as versatile platforms for gene regulation. Curr Opin Genet Dev 2014;25:110-7.
- Capelson M, Hetzer M W. The role of nuclear pores in gene regulation, development and disease. EMBO Rep 2009;10:697-705.
- Bustamante JO, Michelette ER, Geibel JP, Dean DA, Hanover JA, McDonnell TJ. Calcium, ATP and nuclear pore channel gating. Pflügers Arch 2000;439:433-44.
- Walther T C, Askjaer P, Gentzel M, et al. RanGTP mediates nuclear pore complex assembly. Nature. 2003;424:689-94.
- Harel A, Chan R C, Lachish-Zalait A, Zimmerman E, Elbaum M, Forbes D J. Importin β negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol Biol Cell 2003;14:4387-96.
- Cordes VC, Rackwitz HR, Reidenbach S. Mediators of nuclear protein import target karyophilic proteins to pore complexes of cytoplasmic annulate lamellae. Exp Cell Res 1997;237:419-33.
- Imreh G, Hallberg E. An integral membrane protein from the nuclear pore complex is also present in the annulate lamellae: implications for annulate lamellae formation. Exp Cell Res 2000;259:180-90.
- Ewald A, Kossner U, Scheer U, Dabauvalle MC. A biochemical and immunological comparison of nuclear and cytoplasmic pore complexes. J Cell Sci 1996;109:1813-24.
- Belgareh N, Doye V. Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells. J Cell Biol 1997;136:747-59.
- Toyama BH, Savas JN, Park SK, et al. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 2013;154:971-82.
- Favreau C, Bastos R, Cartaud J, Courvalin J C, Mustonen P. Biochemical characterization of nuclear pore complex protein gp210. Eur J Biochem. 2001;268:3883-9.
- Kihlmark M, Imreh G, Hallberg E. Sequential degradation of proteins from the nuclear envelope during apoptosis. J Cell Sci 2001;114:3643-3653.
- Lenz-Böhme B, Wismar J, Fuchs S, et al. Insertional mutation of the Drosophila nuclear lamin Dm0 gene results in defective nuclear envelopes, clustering of nuclear pore complexes, and accumulation of annulate lamellae. J Cell Biol 1997;137:1001-16.
- Heath CV, Copeland CS, Amberg DC, Priore VD, Snyder M, Cole CN. Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutation of the Saccharomyces cerevisiae RAT2/NUP120 gene. J Cell Biol 1995;131:1677-97.
- Aitchison JD, Blobel G, Rout MP. Nup120p: A yeast nucleoporin required for NPC distribution and mRNA transport. J Cell Biol 1995;131:1659-75.
- Pemberton L, Rout MP, Blobel G. Disruption of the nucleoporin gene NUP133 results in clustering of nuclear pore complexes. Proc Natl Acad Sci USA 1995;92:1187-91.
- Maul GD, Deaven L. Quantitative determination of nuclear pore complexes in cycling cells with differing DNA content. J Cell Biol 1977;73:748-60.
- Maul GG, Deaven LL, Freed JJ, Campbell GL, Beçak W. Investigation of the determinants of nuclear pore number. Cytogenet Cell Genet 1980;26:175-90.
- Maul HM, Hsu BY, Borun TM, Maul GG. Effect of metabolic inhibitors on nuclear pore formation during the HeLa S3 cycle. J Cell Biol 1973;59:669-76.
- Iouk T, Kerscher O, Scott R J, Basrai MA, Wozniak RW. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J Cell Biol 2002;159:807-19.
- Salina D, Enarson P, Rattner J B, Burke B. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J Cell Biol 2003;162:991-1001.
- Stukenberg P T, Macara I G. The kinetochore NUPtials. Nat Cell Biol 2003;5:945-7.
- Rout MP, Wente SR. Pores for thought: Nuclear pore complex proteins. Trends Cell Biol 1994;4:357-65.
- Wu X, Kasper LH, Mantcheva RT, Mantchev GT, Springett MJ, vanDeursen JM. Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function. Proc Natl Acad Sci USA 2001;98:3191-6.
- Powers MA, Forbes DJ, Dahlberg JE, Lund E. The vertebrate GLFG nucleoporin, Nup98, is an essential component of multiple RNA export pathways. J Cell Biol 1997;136:241-50.
- Bayliss R, Littlewood T, Stewart M. Structural basis for the interaction between FxFG nucleoporin repeats and importin-β in nuclear trafficking. Cell 2000;102:99-108.
- Ando D, Zandi R, Kim Y W, Colvin M, Rexach M, Gopinathan A. Nuclear pore complex protein sequences determine overall copolymer brush structure and function. Biophys J 2014;1997-2007.
- Lim RY, Fahrenkrog B, Koser J, Schwarz-Herion K, Deng J, Aebi U. Nanomechanical basis of selective gating by the nuclear pore complex. Science 2007;318:640-3.
- Ribbeck K, Görlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J 2001;20:1320-30.
- Frey S, Richter RP, Gorlich D. FS-rich repeats of nuclear pore proteins from a three-dimensional mesh work with hydrogel-like properties. Science 2006;314:815-7.
- Yamada J, Phillips JL, Patel S, et al. A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol Cell Proteomics 2010;9:2205-24.
- Izaurralde E, Kutay U, vonKobbe C, Mattaj I W, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J 1997;16:6535-47.
- Pu RT, Dasso M. The balance of RanBP1 and RCC1 is critical for nuclear assembly and nuclear transport. Mol Biol Cell 1997;8:1955-70.
- Avis JM, Clarke PR. Ran, a GTPase involved in nuclear processes: Its regulators and effectors. J Cell Sci 1996;109:2423-7.
- Azuma Y, Dasso M. The role of Ran in nuclear function. Curr Opin Cell Biol 2000;12:302-7.
- Nachury MV, Weis K. The direction of transport through the nuclear pore can be inverted. Proc Natl Acad Sci USA 1999;96:9622-7.
- Ben-Efraim I, Gerace L. Gradient of increasing affinity of importin beta for nucleoporins along the pathway of nuclear import. J Cell Biol 2001;152:411-7.
- Kehlenbach R H, Dickmanns A, Kehlenbach A, Guan T, Gerace L. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J Cell Biol 1999;145:645-57.
- Singh BB, Patel HH, Roepman R, Schick D, Ferreira P A. The zinc finger cluster domain of RanBP2 is a specific docking site for the nuclear export factor, exportin-1. J Biol Chem 1999;274:37370-37378.
- Shah S, Forbes DJ. Separate nuclear import pathways converge on the nucleoporin Nup153 and can be dissected with dominant- negative inhibitors. Curr Biol 1998;8:1376-86.
- Ullman KS, Shah S, Powers MA, Forbes DJ. The nucleoporin nup153 plays a critical role in multiple types of nuclear export. Mol Biol Cell 1999;10:649-64.
- Kapinos LE, Schoch RL, Wagner RS, Schleicher KD, Lim RY. Karyopherin-centric control of nuclear pores based on molecular occupancy and kinetic analysis of multivalent binding with FG nucleoporins. Biophys J 2014;106:1751-62.
- Schoch RL, Kapinos LE, Lim RY. Nuclear transport receptor binding avidity triggers a self-healing collapse transition in FG-nucleoporin molecular brushes. Proc Natl Acad Sci USA 2012;109:16911-6.
- Zwerger M, Eibauer M, Medalia O. Insights into the gate and scaffold of the nuclear pore complex. Nucleus 2016;7:1-7.
- Tu LC, Musser SM. Single molecule studies of nucleocytoplasmic transport. Biochem Biophys Acta 20111813:1607-18.
- Kalderon D, Richardson WD, Markham AF, Smith A E. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 1984;311:33-8.
- Dingwall C, Sharnick SV, Laskey RA. A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell 1984;30:449-58.
- Dingwall C, Dilworth SM, Black SJ, Kearsey SE, Cox LS, Laskey RA. Nucleoplasmin cDNA sequence reveals polyglutamic acid tracts and a cluster of sequences homologous to putative nuclear localization signals. EMBO J 1987;6:69-74.
- Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol 1995;129:551-60.
- Henderson B R, Eleftheriou A. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp Cell Res 2000;256:213-224.
- Cullen BR. Nuclear RNA export. J Cell Sci 2003;116:587-97.
- Zhu J, Shibasaki F, Price R, et al. Intramolecular masking of nuclear import signal on NF-AT4 by casein kinase I and MEKK1. Cell 1998;93:851-61.
- Okamura H, Aramburu J, Garcia-Rodriguez C, et al. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol Cell 2000;6:539-50.
- Hay RT, Vuillard L, Desterro JM, Rodriguez MS. Control of NF-kappa B transcriptional activation by signal induced proteolysis of I kappa B alpha. Philos Trans R Soc Lond B Biol Sci 1999;354:1601-9.
- Rihs HP, Jans DA, Fan H, Peters R. The rate of nuclear cytoplasmic protein transport is determined by the casein kinase II site flanking the nuclear localization sequence of the SV40 T-antigen. EMBO J 1991;10:633-9.
- Matunis MJ, Wu J, Blobel G. SUMO-1 modification and its role in targeting the RanGTPase-activating protein, RanGAP1, to the nuclear pore complex. J Cell Biol 1998;140:499-509.
- Melchior F, Schergaut M, Pichler A. SUMO: Ligases, isopeptidases and nuclear pores. Trends Biochem Sci 2003;28:612-8.
- Tasken K, Aandahl EM. Localized effects of cAMP mediated by distinct routes of protein kinase A. Physiol Rev 2004;84:137-67.
- Feil R, Lohmann S M, deJonge H, Walter U, Hofmann F. Cyclic GMP-dependent protein kinases and the cardiovascular system: insights from genetically modified mice. Circ Res 2003;93:907-16.
- Schlossmann J, Feil R, Hofmann F. Signaling through NO and cGMP-dependent protein kinases. Ann Med 2003;35:21-7.
- Heist E K, Schulman H. The role of Ca2+/calmodulin-dependent protein kinases within the nucleus. Cell Calcium. 1998;23:103-14.
- Sugiura M, Kono K, Liu H, Shimizugawa T, Minekura H, Spiegel S, Kohama T. Ceramide kinase, a novel lipid kinase. Molecular cloning and functional characterization. J Biol Chem 2002;277:23294-300.
- Kanoh H ,Yamada K, Sakane F. Diacylglycerol kinases: emerging downstream regulators in cell signaling systems. J Biochem (Tokyo) 2002;131:629-33.
- Chahine MN, Blackwood DP, Dibrov E, et al. Oxidized low density lipoprotein affects smooth muscle cell growth through MAPK- medicated actions on nuclear protein import. J Mol Cell Cardiol 2009;46:431-41.
- Klumpp S, Krieglstein J. Phosphorylation and dephosphorylation of histidine residues in proteins. Eur J Biochem 2002;269:1067-71.
- Khokhlatchev AV, Canagarajah B, Wilsbacher J, et al. Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell 1998;93:605-15.
- Xiao C.-Y, Hübner S, Jans D A. SV40 large tumor antigen nuclear import is regulated by the double-stranded DNA-dependent protein kinase site (serine 120) flanking the nuclear localization sequence. J Biol Chem 1997;272:22191-8.
- Barford D, Das AK, Egloff MP. The structure and mechanism of protein phosphatases: Insights into catalysis and regulation. Annu Rev Biophys Biomol Struct 1998;27:133-64.
- Tonks N K, Cohen P. Calcineurin is a calcium ion-dependent, calmodulin-stimulated protein phosphatase. Biochim Biophys Acta 1983;747:191-3.
- Bollen M, Beullens M. Signaling by protein phosphatases in the nucleus. Trends Cell Biol 2002;12:138-45.
- Lubert EJ, Sarge KD. Interaction between protein phosphatase 2A and members of the importin β superfamily. Biochem Biophys Res Comm 2003;303:908-13.
- Faustino RS, Maddaford, TG, Pierce GN. Mitogen activated protein kinase at the nuclear pore complex. J Cell Mol Med 2011;15:928-37.
- Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607-60.
- Adam EJ, Adam SA. Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J Cell Biol 1994;125:547-55.
- Chi NC, Adam EJ, Adam SA. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J Cell Biol 1995;130:265-74.
- Cingolani G, Petosa C, Weis K, Müller C W. Structure of importin-β bound to the IBB domain of importin-α. Nature 1999;399:221-9.
- Kutay U, Izaurralde E, Bischoff FR, Mattaj I W, Görlich D. Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex. EMBO J 1997;16:1153-63.
- Chi NC, Adam SA. Functional domains in nuclear import factor p97 for binding the nuclear localization sequence receptor and the nuclear pore. Mol Biol Cell 1997;8:945-56.
- Chi NC, Adam EJ, Adam SA. Different binding domains for Ran- GTP and Ran-GDP/RanBP1 on nuclear import factor p97. J Biol Chem 1997;272:6818-22.
- Adam SA, Gerace L. Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell 1991;66:837-47.
- Cingolani G, Lashuel HA, Gerace L, Müller CW. Nuclear import factors importin α and importin β undergo mutually induced conformational changes upon association. FEBS Lett 2000;484:291-8.
- Palmeri D, Malim MH. Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol Cell Biol 1999;19.
- Henderson BR, Percipalle P. Interactions between HIV Rev and nuclear import and export factors: The Rev nuclear localisation signal mediates specific binding to human importin-beta. J Mol Biol 1997;274.
- Truant R, Cullen BR. The arginine-rich domains present in human immunodeficiency virus type 1 tat and rev function as direct importin beta-dependent nuclear localization signals. Mol Cell Biol 1999;19.
- Jäkel S, Görlich D. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J 1998;17:4491-502.
- Lam MH, Briggs LJ, Hu W, Martin TJ, Gillespie M T, Jans D A. Importin β recognizes parathyroid hormone-related protein with high affinity and mediates its nuclear import in the absence of importin α. J Biol Chem 1999;274:7391-8.
- Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell 1998;94:193-204.
- Fontes M R M, Teh T, Kobe B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α. J Mol Biol 2000;297:1173-94.
- Melén K, Fagerlund R, Franke J, Köhler M, Kinnunen L, Julkunnen I. Importin α nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J Biol Chem 2003;278:28193-200.
- Hood J K, Silver P A. Cse1p is required for export of Srp1p/ importin-α from the nucleus in Saccharomyces cerevisiae. J Biol Chem 1998;273:35142-6.
- Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell 1997;90:1061-71.
- Herold A, Truant R, Wiegand H, Cullen BR. Determination of the functional domain organization of the importin α nuclear import factor. J Cell Biol 1998;143:309-18.
- Kobe B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin α. Nat Struct Biol 1999;6:388-97.
- Harreman MT, Hodel MR, Fanara P, Hodel AE, Corbett AH. The auto-inhibitory function of importin α is essential in vivo. J Biol Chem. 2003;278:5854-5863.
- Köhler M, Speck C, Christiansen M, et al. Evidence for distinct substrate specificities of importin α family members in nuclear protein import. Mol Cell Biol 1999;19:7782-91.
- Kamei Y, Yuba S, Nakayama T, Yoneda Y. Three distinct classes of the α-subunit of the nuclear pore-targeting complex (importin-α) are differentially expressed in adult mouse tissues. J Histochem Cytochem 1999;47:363-72.
- Talcott B, Moore MS. The nuclear import of RCC1 requires a specific nuclear localization sequence receptor, karyopherin α3/Qip. J Biol Chem 2000;275:10099-104.
- Huber J, Cronshagen U, Kadokura M, et al. Snurportin1, an m3G- cap-specific nuclear import receptor with a novel domain structure. EMBO J 1998;17:4114-26.
- Huber J, Dickmanns A, Lührmann R. The importin-β binding domain of snurportin1 is responsible for the Ran- and energy- independent nuclear import of spliceosomal U snRNPs in vitro. J Cell Biol 2002;156:467-79.
- Belmont A. Dynamics of chromatin, proteins, and bodies within the cell nucleus. Curr Opin Cell Biol 2003;15:304-10.
- Bäuerle M, Doenecke D, Albig W. The requirement of H1 histones for a heterodimeric nuclear import receptor. J Biol Chem 2002;277:32480-9.
- Brown DT. Histone H1 and the dynamic regulation of chromatin function. Biochem Cell Biol 2003;81:221-7.
- Jäkel S, Albig W, Kutay U, et al. The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J 1999;18:2411-23.
- Jullien D, Görlich D, Laemmli UK, Adachi Y. Nuclear import of RPA in Xenopus egg extracts requires a novel protein XRIPα but not importin α. EMBO J 1999;18:4348-58.
- Wold MS. Replication protein A: Heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem 1997;66:61-92.
- dachi Y, Laemmli UK. Study of the cell cycle-dependent assembly of the DNA pre-replication centers in Xenopus egg extracts. EMBO J 1994;13:4153-64.
- Moore SP, Erdile L, Kelly T, Fishel R. The human homologous pairing protein HPP-1 is specifically stimulated by the cognate single-stranded binding protein hRP-A. Proc Natl Acad Sci USA 1991;88:9067-71.
- Shinohara A, Shinohara M, Ohta T, Matsuda S. Ogawa T. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells 1998;3:145-56.
- Longhese MP, Plevani P, Lucchini G. Replication factor A is required in vivo for DNA replication, repair and recombination. Mol Cell Biol 1994;14:7884-90.
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 1998;391:407-10.
- Fried H M, Kutay U. Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci 2003;60:1659-88.
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 1997;90:1051-60.
- Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell 1995;82:463-73.
- Fischer U, Huber J, Boelens WC, Mattaj I W, Lührmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 1995;82:475-83.
- Bischoff F R, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. RanGAP1 induces GTPase activity of nuclear ras-related Ran. Proc Natl Acad Sci USA 1994;91:2587-91.
- Scherf U, Kalab P, Dasso M, Pastan I, Brinkmann U. The hCSE1/ CAS protein is phosphorylated by HeLa extracts and MEK-1: MEK-1 phosphorylation may modulate the intracelluar localization of CAS. Biochem Biophys Res Comm 1998;250:623-8.
- Scherf U, Pastan I, Willingham M C, Brinkmann U. The human CAS protein which is homologous to the CSE1 yeast chromosome segregation gene product is associated with microtubules and mitotic spindle. Proc Natl Acad Sci USA 1996;93:2670-4.
- Ogryzko V V, Brinkmann E, Howard B H, Pastan I, Brinkmann U. Antisense inhibition of CAS, the human homologue of the yeast chromosome segregation gene CSE1, interferes with mitosis in HeLa cells. Biochemistry. 1997;36:9493-9500.
- Faustino RS, Cheung P, Richard MN, et al. Ceramide regulation of nuclear protein import. J Lipid Res 2008;49:654-662
- Holaska JM, Black BE , Love D C, Hanover JA, Leszyk J, Paschal BM. Calreticulin is a receptor for nuclear export. J Cell Biol 2001;152:127-40.
- Mesaeli N, Nakamura K, Zvaritch E, et al. Calreticulin is essential for cardiac development. J Cell Biol 1999;144:857-68.
- Li J, Pucéat M, Perez-Terzic C, et al. Calreticulin reveals a critical Ca2+ checkpoint in cardiac myofibrillogenesis. J Cell Biol 2002;158:103-13.
- Walther RE, Lamprecht C, Ridsdale A, et al. Nuclear export of the glucocorticoid receptor is accelerated by cell fusion-dependent release of calreticulin. J Biol Chem 2003;278:37858-64.
- DeFranco D B. Nuclear export: DNA-binding domains find a surprising partner. Curr Biol 2001;11:R1036-R1037.
- Holaska J M, Black B E, Rastinejad F, Paschal B M. Ca2+-dependent nuclear export mediated by calreticulin. Mol Cell Biol 2002;22:6286-97.
- Michalak M, Corbett E F, Mesaeli N, Nakamura K, Opas M. Calreticulin: One protein, one gene, many functions. Biochem J 1999;344:281-92.
- Arts GJ, Kuersten S, Romby P, Ehresmann B, Mattaj I W. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J 1998;17:7430-41.
- Kutay U, Lipowsky G, Izaurralde E, et al. Identification of a tRNA- specific nuclear export receptor. Mol Cell 1998;1:359-69.
- Calado A, Treichel N, Müller EC, Otto A, Kutay U. Exportin-5- mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J 2002;21:6216-24.
- Schmitt I, Gerace L. In vitro analysis of nuclear transport mediated by the C-terminal shuttle domain of Tap. J Biol Chem 2001;276.
- Lévesque L, Guzik B, Guan T, et al. RNA export mediated by Tap involves NXT1-dependent interactions with the nuclear pore complex. J Biol Chem 2001;276:44953-62.
- Guzik BW, Levesque L, Prasad S, et al. NXT1 (p15) is a crucial cellular cofactor in TAP-dependent export of intron-containing RNA in mammalian cells. Mol Cell Biol 2001;21:2545-54.
- Wiegand HL, Coburn GA, Zeng Y, Kang Y, Bogerd HP, Cullen BR. Formation of Tap/NXT1 heterodimers activates Tap-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes. Mol Cell Biol 2002;22:245-56.
- Gilbert W, Guthrie C. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol Cell 2004;13:201-212.
- Hurt E, Luo M J, Rother S, Reed R, Strasser K. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc Natl Acad Sci USA 2004;101:1858-62.
- Dean KA, vonAhsen O, Görlich D, Fried HM. Signal recognition particle protein 19 is imported into the nucleus by importin 8 (RanBP8) and transportin. J Cell Sci 2001;114:3479-85.
- Kehlenbach R H, Gerace L. Phosphorylation of the nuclear transport machinery down-regulates nuclear protein import in vitro. J Biol Chem. 2000;275:17848-56.
- Stochaj U, Rother KL. Nucleocytoplasmic trafficking of proteins: with or without Ran? Bioessays 1999;21:579-589.
- Melchior F, Paschal B, Evans J, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol 1993;123:1649-59.
- Quimby BB, Lamitina T, L’Hernault SW, Corbett AH. The mechanism of Ran import into the nucleus by nuclear transport factor 2. J Biol Chem 2000;275:28575-28582.
- Yoneda Y, Hieda M, Nagoshi E, Miyamoto Y. Nucleocytoplasmic protein transport and recycling of Ran. Cell Struct Funct 1999;24:425-33.
- Richards SA, Carey KL, Macara IG. Requirement of guanosine triphosphate-bound Ran for signal-mediated nuclear protein export. Science 1997;276:1842-4.
- Corbett AH, Koepp DM, Schlenstedt G, Lee MS, Hopper AK, Silver PA. Rna1p, a Ran/TC4 GTPase activating protein, is required for nuclear import. J Cell Biol 1995;130:1017-26.
- Nehrbass U, Blobel G. Role of the nuclear transport factor p10 in nuclear import. Science 1996;272:120-2.
- Chaillan-Huntington C, Braslavsky CV, Kuhlmann J, Stewart M. Dissecting the interactions between NTF2, RanGDP, and the nucleoporin XFXFG repeats. J Biol Chem 2000;275:5874-9.
- Bayliss R, Ribbeck K, Akin D, et al. Interaction between NTF2 and xFxFG-containing nucleoporins is required to mediate nuclear import of RanGDP. J Mol Biol 1999;293:579-93.
- Ribbeck K, Lipowsky G, Kent HM, Stewart M, Görlich D. NTF2 mediates nuclear import of Ran. EMBO J 1998;17:6587-98.
- Smith A, Brownawell A, Macara I G. Nuclear import of Ran is mediated by the transport factor NTF2. Curr Biol 1998;8:1403-6.
- Steggerda SM, Black BE, Paschal BM. Monoclonal antibodies to NTF2 inhibit nuclear protein import by preventing nuclear translocation of the GTPase Ran. Mol Biol Cell 2000;11:703-19.
- Geyer M, Assheuer R, Klebe C, et al. Conformational states of the nuclear GTP-binding protein Ran and its complexes with the exchange factor RCC1 and the effector protein RanBP1. Biochemistry 1999;38:11250-60.
- Ohtsubo M, Okazaki H, Nishimoto T. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J Cell Biol 1989;109:1389-97.
- Klebe C, Bischoff FR, Ponstingl H, Wittinghofer A. Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry 1995;34:639-47.
- Yoneda Y. Nucleocytoplasmic protein traffic and its significance to cell function. Genes Cells 2000;5:777-787.
- Moroianu J, Blobel G. Protein export from the nucleus requires the GTPase Ran and GTP hydrolysis. Proc Natl Acad Sci USA 1995;92:4318-22.
- Nemergut ME, Macara IG. Nuclear import of Ran exchange factor, RCC1, is mediated by at least two distinct mechanisms. J Cell Biol 2000;149:835-49.
- Lounsbury KM, Macara IG. Ran-binding protein 1 (RanBP1) forms a ternary complex with Ran and karyopherin β and reduces Ran GTPase-activating protein (RanGAP) inhibition by karyopherin β. J Biol Chem 1997;272:551-5.
- Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science 2002;295:2452-6.
- Takizawa CG, Weis K, Morgan DO. Ran-independent nuclear import of cyclin B1-Cdc2 by importin β. Proc Natl Acad Sci USA 1999;96:7938-43.
- Ribbeck K, Kutay U, Paraskeva E, Görlich D. The translocation of transportin-cargo complexes through nuclear pores is independent of both Ran and energy. Curr Biol 1999;9:47-50.
- Sachdev S, Bagchi S, Zhang DD, Mings AC, Hannink M. Nuclear import of IκBα is accomplished by a Ran-independent transport pathway. Mol Cell Biol 2000;20:1571-82.
- Yokoya F, Imamoto N, Tachibana T, Yoneda Y. β-catenin can be transported into the nucleus in a Ran-unassisted manner. Mol Biol Cell 1999;10:1119-31.
- Hetzer M, Mattaj IW. An ATP-dependent, Ran-independent mechanism for nuclear import of the U1A and U2B'' spliceosome proteins. J Cell Biol 2000;148:293-303.
- Faustino RS, Stronger LN, Landry MN, et al. Lysophosphatidylcholine stimulates nuclear protein import in vascular smooth muscle cells via stimulation of RanGAP activity. Mol Pharmacol 2007;71:438-45.
- Richard MN, Deniset JF, Kneesch AL, et al. Mechanical stretch stimulates smooth muscle cell growth, nuclear protein import and nuclear pore expression through mitogen-activated protein kinase activation. J Biol Chem 2007;282:23081-8.
- Czubryt MP, Austria JA, Pierce GN. Hydrogen peroxide inhibition of nuclear protein import in aortic vascular smooth muscle cells is mediated by the MAP kinase ERK2. J Cell Biol 2000;148:7-15.
- Zettler M E, Prociuk M A, Austria J A, et al. Oxidized low density lipoprotein retards the growth of proliferating cells by inhibiting nuclear translocation of cell cycle proteins. Arterioscl Thromb Vasc Biol 2004;24:1-7.
- Chahine MN, Dibrov E, Blackwood DP, et al. Oxidized LDL enhances stretch-induced smooth muscle cell proliferation through alterations in nuclear protein import. Can J Physiol Pharmacol 2012;90:1559-68.
- Chahine MN, Mioulane M, Sikkel M B, et al. Nuclear pore rearrangements and nuclear trafficking in cardiomyocytes from rat and human failing hearts. Cardiovasc Res 2015;105:31-43.
- Chahine M N, Pierce G N. Therapeutic targeting of nuclear protein import in pathological cell conditions. Pharmacol Rev 2009;61:358-72.
- *Corresponding Author:
- Dr Grant N Pierce
Institute of Cardiovascular Sciences, Albrechtsen Research Centre, St Boniface Hospital, 351 Tache Avenue, Winnipeg, Manitoba R2H 2A6.
Telephone: 204-235-3206
Fax: 204-235-0793
E-mail: gpierce@sbrc.ca
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact support@pulsus.com
Abstract
Nucleocytoplasmic trafficking describes the process whereby molecules destined for the nucleoplasm or cytoplasm move across the nuclear envelope. This transport regulates transcription and translation by respective control of transnuclear movements of transcription factors and messenger RNA. Transport across the nuclear envelope occurs through large MDa pores embedded in the inner and outer nuclear membranes, and is a dynamic, adaptable process modulated by biochemical and biophysical cellular milieus. The present review discusses current understanding of the composition and function of the nuclear pore complex through which transport occurs; regulatory elements within molecular cargo that influence its movement into and out of the nucleus; transport associated proteins that guide the cargo across the nuclear envelope; and factors responsible for modulating nucleocytoplasmic transport.
-Keywords
Nuclear pore complex; Nuclear transport
Nucleocytoplasmic Trafficking
Nuclear transport is a complex process involving soluble, cytosolic machinery and membrane-bound components. Molecules that localize from cytosolic to nucleoplasmic compartments and vice versa must cross both the outer and inner nuclear membranes of the nuclear envelope (NE) to reach their destination. For this purpose, a large multiproteinaceous structure known as the nuclear pore complex (NPC) spans both membranes of the NE and serves as the gateway through which molecules transit [1-10].
The NPC
Structure and composition
Electron microscopy of an NE in cross-section demonstrates how the NPC serves as a proteinaceous ‘rivet’ connecting both outer and inner nuclear membranes of the NE. Immunocytochemistry using mAb414, an antibody that recognizes NPC proteins or nucleoporins (nups), demonstrates intense staining of the nucleus as well as cytoplasmic annulate lamellae (Figure 1). Confocal microscopy of smooth muscle cells illustrates regular distribution of NPCs across the surface of the NE. The NPC is a dynamic and highly regulated structure, which shows a high degree of evolutionary conservation across various species [7]. Most of the information on NPC composition and architecture has been garnered from yeast [10] and Xenopus [11,12]; however, the primary focus of this section will be on mammalian NPCs.
Figure 1: Visualization of a vascular smooth muscle cell nucleus using confocal microscopy. Images of mAb414-stained nuclei were obtained via confocal microscopy at 100× magnification. Punctate staining of the nuclear envelope is immediately apparent, which is a distinctive nuclear pore complex staining pattern. Cytosolic staining of pore proteins within annulate lamellae is also apparent
Mammalian NPCs are approximately 125 MDa and composed of 30 to 32 discrete nups that occur in redundant groupings within the pore [7]. Overall, the NPC is approximately 120 nm in diameter and possesses an axial depth of approximately 70 nm. It is composed of eight multimeric subunits arranged in an annular configuration that can be visualized as a distinctive barrel-like shape. This structural conformation leaves a functionally dynamic aqueous central channel approximately 9 nm to 11 nm in diameter under normal conditions [13,14]. Electron microscopy of prepared NEs illustrates the octagonal symmetry of the pore when viewed en face. The central pore can dilate to accommodate larger cargoes in transit [15,16] as well as respond to calcium fluxes [17,18]. In addition to the main central channel [19,20], peripheral channels exist, which are hypothesized to permit the flow of various small molecules and ions [4,21]. One feature of the NPC core that remains controversial is the pore plug or central transporter. In some preparations, electron and atomic force microscopy have revealed the presence of a large molecule occluding the central channel of the pore [1]. It is unclear whether this plug is a bonafide structural component of the pore or simply trapped cargo; however, observations made by Stoffler et al [22] and others [23,24] suggest that the central plug may be a composite of a molecule caught in transit with attendant changes in distal ring conformation.
On the nuclear and cytoplasmic faces of the pore are ring structures from which filaments extend into their respective compartments [4]. The cytoplasmic and nuclear ring-filament assemblies are asymmetrical and possess distinct structural variations. The cytoplasmic filaments are regularly spaced around the ring and extend into the cytosol for approximately 50 nm [22]. They also contain nups, which serve as docking sites for pretransport import complexes [25-28].
The nuclear filaments are evenly distributed around the nuclear ring, which in turn interacts with the nuclear lamina [29]. The nuclear filaments extend significantly further into the nucleoplasm than their cytoplasmic counterparts and are joined at their distal ends by a smaller ring, termed the distal ring [3,4,22,29]. This entire assembly is known as the ‘nuclear basket’ [3,4]. Sites on the distal ring and at the distal ends of the nuclear filaments serve as initial docking sites for export complexes and serve a function similar to their cytosolic counterparts [30]. Another unique feature of the nuclear face of the NPC is its association with the nuclear lamina (via the nuclear ring) [29]. The function of the nuclear lamina in this respect is to anchor the NPCs within the NE [31].
Dynamics
The nuclear pore is a dynamic structure that responds to a variety of stimuli [32-35], and its functional diversity extends beyond its role as a transport gate, discussed in excellent reviews elsewhere [36,37]. The presence or absence of the central transporter in response to the depletion or repletion of periplasmic calcium stores, respectively, is an excellent example of NPC dynamics [17,18]. It has been proposed that this transporter – or ‘plug’ – may represent a calcium-sensitive multimer composed of nup gp210 [32]. Others have suggested that this feature is a preparative artifact because it was not observed in other experiments and was postulated to be molecular cargo trapped in transit (35). Numerous studies investigating the role of calcium in NPC regulation, however, have demonstrated a calcium-sensitive mechanism responsible for closing and opening the pore [17,18]. These independent observations support the notion that the NPC is dynamically regulated by calcium fluxes in and around the nucleus [17,18,38].
Nups are capable of diffusing from one membrane system to another within the same cell. Partial pore complexes and individual nups have been localized to the cytoplasmic annulate lamellae (AL), an organelle associated with components of the Ran cycle and potentially involved in nup distribution [39-41]. Morphologically, the AL are multilayered membrane structures [31,41,42]. Harel et al [40] demonstrated that imbalances in cellular RanGTP concentrations lead to excessive formation of de novo AL; therefore, the presence of AL may be a secondary effect of nuclear assembly processes. However, the identification of partial pore complexes within the AL suggests that the AL may serve as a distributor and repository for pre-assembly NPC proteins and nuclear pore subcomplexes [41]. This was based on the observation that immunofluorescent staining for nups visualized proteins within the NPC as well as within the AL [43]. Belgareh and Doye [44] reported transnuclear diffusion of a nuclear pore protein in a binucleate heterokaryon, demonstrating that some nups are capable of translocating from one membrane system to another. This finding provides evidence of the hypothetical role of the AL as a nup repository and supports the dynamic characteristic of the NPC. Interphasic NPCs have low turnover in living cells and are replaced after a round of mitosis [31]. Significantly, individual nups have variable half-lives [45], with some nups demonstrating off-pore mobility [31,46]. In untreated cells, individual NPCs possess limited lateral movement and show heterogeneous distribution because they are anchored to the nuclear lamina via the nuclear ring [31]. They are capable, however, of shifting in waves around the entire surface of the nucleus as a single entity [31], which can be envisioned as a net covering a ball (ie, although the entire net can freely move around the surface of the ball, the interconnecting points are not significantly displaced with respect to one another). Abnormal spatial distribution of NPCs has been reported in conjunction with alterations or defects in nuclear lamins [47,48]. Mutations in a specific subset of nups can also result in NPC clustering instead of the normal distribution [44,49-51]. These observations imply that, although NPCs maintain specific distributions under normal conditions, they are capable of collectively undergoing significant spatial rearrangement and are not static structures. Additionally, the density of NPCs can change according to the metabolic needs of the cell [52-54]. Changes in the nuclear pore can, thus, be correlated with growth requirements and/or changes in mitosis.
The link between NPC dynamics and mitosis has been further strengthened by work that has demonstrated the redistribution of integral nups to the kinetochore during mitosis [55-57]. The trigger responsible for relocalizing these NPC proteins has been presumed to be NE breakdown [57], which releases the nups, enabling them to associate with the mitotic machinery [57]. The cell can then proceed past the spindle checkpoint and subsequent steps in mitosis can continue unimpeded. From these observations, it is clear that individual nuclear pore proteins are mobile in both interphase and mitosis, significantly more so in the latter.
Models of Transport
Despite our present knowledge of the structure and characteristics of the nuclear pore, the nature of the barrier that regulates passage of molecules across the NPC remains to be clearly defined. Early research in yeast demonstrated that some nups contained FG repeats as well as GLFG and FxFG motifs [58]. These sequences are highly conserved between NPCs of different species and facilitate searches for homologous proteins in higher eukaryotic species. Importantly, the interaction of nups with soluble components of the nuclear transport machinery occurs through these FG repeats [59-61] and forms the basis of several models of nuclear transport.
Polymer brush
A thorough analysis of the yeast NPC by Rout et al [10] revealed that the yeast NPC contains approximately 30 to 32 nups. Based on the number and distribution of nups observed in their work, Rout et al [10] suggested Brownian affinity as a potential mechanism of transport. The notion of transport here was that import or export complexes bind to FG nups within the NPC, increasing their residence time at the pore opening, thus raising the probability of their entry into the channel and subsequent translocation through the central channel by Brownian motion [10]. This initial understanding served as the conceptual precursor to the present polymer brush model currently proposed
In this model, disordered FG domains extend to create a virtual gate, or entropic brush region that blocks non-NLS-mediated protein movement [62]. In contrast, binding via importin-b collapsed the extended domain. Furthermore, this effect was reversed by the action of RanGTP that would return the collapsed regions to their extended, brush conformation [63].
hydrogel
Another model that attempts to take into account cargo movement is the hydrogel model [64,65]. This model hypothesizes the existence of an interconnected meshwork between nups that are connected to one another via their FG repeats within the channel [64]. Transport cargoes are able to traverse this meshwork because they are able to interact with the FG repeats themselves, thus breaking the connection between nups within the mesh. Additionally, the ‘holes’ within the mesh act as a molecular sieve and together with the hydrophobic nature of the channel serve to exclude both inert (receptor-less) cargo and molecules beyond the size limit of the pore [64].
Forest (trees and bushes) model
A third proposed mechanism of action focuses on the bimodal distribution of two categories of extended FG conformations and their arrangement within the NPC (66). In this model, FG nups with extended disordered domains (trees) or relatively compact FG regions (bushes) are arranged in a manner that creates distinct transport zones within the central channel. This combines aspects of the polymer brush and hydrogel models, and takes into account the varying affinities of discrete nups for specific cargo-receptor complexes.
The three models described above explain the movement of cargoes across the NE; however, the role of the Ran gradient is not fully considered for each. The polymeric brush model does provide evidence for the role of RanGTP in altering permeability, but it is a fourth model that attempts to explain the selective permeability barrier of the NPC in the context of a full Ran gradient. This is critical to nuclear transport processes, as collapse of the gradient can stop transport [67-70]. Additionally, the direction of transport can be reversed in the presence of high cytosolic concentrations of RanGTP [71].
Karyopherin-centric, binding affinity model
Ben-Efraim and Gerace proposed a model that would incorporate the directionality and motive force provided by RanGTP/GDP distribution across the NE with size exclusion limits and binding affinities [72]. In their model, transport cargoes bound to RanGTP move up an increasing affinity gradient to end up at terminal docking sites in their predestined compartment [72-76]. This model reconciled biophysical parameters of the NPC with the observed transport dynamics, and would ultimately serve as the conceptual precursor of the contemporary karyopherin-centric model [77,78]. This model incorporates the observation that specific FG-containing nups bind transport receptors with varying degrees of affinity [72].
While all proposed models explain the exclusivity of the selective FG nup gating mechanism to some degree, no consensus has yet been reached that unifies the spectrum of biophysical properties exhibited by the NPC barrier with the diversity of biochemical factors critical for nucleocytoplasmic transport. Future work using technology capable of increased NPC structural detail and high temporospatial resolution [79,80] of discrete transport events will contribute greatly to addressing this long-standing challenge within the field.
Nuclear Transport Signal Sequences
Nuclear localization signals
Nuclear transport generally describes the process by which molecules move back and forth between the nuclear and cytoplasmic spaces. It is a bidirectional and energy-dependent phenomenon, with specific regulatory mechanisms distributed within the soluble portion of the cell as well as at the NE. Nucleocytoplasmic trafficking is initiated on recognition of a specific signal sequence that directs the molecule to either be imported into the nucleus from the cytoplasm or exported in the opposite direction.
The first signal sequence identified was a monopartite, polybasic amino acid motif (PKKKRKV) directly responsible for the nuclear localization of the SV40 large T antigen [81]. This nuclear localization signal (NLS) was a polylysine motif known as the ‘classical’ signal sequence (Table 1). NLSs also exist in a bipartite form. Dingwall et al [82] first characterized the existence of a bipartite NLS in nucleoplasmin and would later demonstrate that it contained polybasic sequences homologous to the NLS of the SV40 large T antigen [83].
| Import receptor | Signal sequence/motif |
|---|---|
| Importin-α | (monopartite) PKKKRKV (3) |
| (bipartite) KRPAAIKKAGQAKKKK (7) | |
| Importin-β | RQARRNRRRRWR (8) |
| Transportin | NQSSNFGPMKGGNFGGRSSGPY |
| GGGGQYFAKPRNQGGY (8) | |
| Snurportin1 | trimethylguanosine cap (m3G) (9) |
| XRIPα | DNA binding domain (10) |
Importin-α is the primary receptor for the ‘classical’ polybasic amino acid motif, which occurs as a monopartite or bipartite signal sequence (see text and above). Once importin-α binds its substrate, an importin-β binding domain on the alpha subunit is exposed, leading to the formation of a heterotrimeric receptor-substrate complex. Importin-β is also a primary receptor and can bind to individual proteins without the need of an adaptor protein. Other import receptors recognize nuclear localization signals distinct from the polylysine type and are depicted above
Table 1: Examples of nuclear import sequences and their cognate receptor
The discovery of the NLS provided the impetus for the identification of other signal sequences responsible for nuclear import, but distinct from those of the SV40 variety. A glycine rich, novel sequence that did not contain any ‘classical’ NLS-type basic amino acids was found in the pre-messenger/mRNA binding protein hnRNP A1 [84] and was designated M9 and recognized by transportin (Table 1). Attachment of this sequence to two cytosolic enzymes, pyruvate kinase and beta-galactosidase, was sufficient to enable their import into the nucleus. A listing of selected NLSs and their cognate receptors are shown in Table 1.
Nuclear export signals
Proteins and RNA exported from the nucleus typically possess a leucinerich signaling motif known as the nuclear export signal (NES). Similar to NLSs, a variety of signaling motifs exist that are capable of mediating nuclear export [85]. Instead of amino acid sequences, RNA species possess structural motifs recognized by specialized exportin molecules that direct them toward the cytoplasm [86]. Examples of various NES are shown (Table 2).
| export receptor | Signal sequence/motif | Molecule exported |
|---|---|---|
| Crm1 | LPPLERLTLD (20) | Rev |
| LALKLAGLDI (21) | PKI | |
| LQKKLEELELE (22) | MEK1 | |
| CAS | CGGLDKIE (25) | importin-α |
| Calreticulin | CGGGKVFFKRAVEGQHNLY (26) | glucocorticoid receptor |
| TAP | RNA stem-loop structure (23) | RNA |
| Mex67p | Polyadenylated RNA (28) | |
Exportins are responsible for delivering proteins to the cytosol from the nucleus. Crm1 (also known as exportin-1) has been well characterized as a nuclear exporter. Other proteins have been identified that bind signals distinctly different from those recognized by Crm1. RNA molecules are exported from the nucleus once their receptor recognizes distinct molecular features (see above)
Table 2: Selected nuclear export signals and their receptors
Due to the variety of transport signals available for import and export, nucleocytoplasmic trafficking remains a tightly controlled process. The primary level of regulation occurs at the nuclear transport signal by controlling access to the NLS/NES. With respect to the localization signal, the eukaryotic cell has evolved various mechanisms to control nuclear trafficking. Phosphorylation, dephosphorylation and regulated proteolysis are some of the common regulatory processes that play a role in regulating nuclear transport at one or more levels [87-92].
Regulation of NlSs and NeSs
A common method of regulation within the eukaryotic cell is phosphorylation of a target substrate. In all phosphorylation and dephosphorylation cycles, there are two classes of enzymes responsible for adding and removing phosphate groups, called kinases and phosphatases, respectively.
A diverse number of kinases exist that can be regulated by cyclic AMP and GMP [93-95], calcium [96] and lipids [97-99]. The process of phosphorylation begins when a protein substrate possesses a motif recognized by the kinase. On association of the kinase with its target, ATP is recruited to the complex and the terminal phosphate (designated as the γ phosphate) is transferred to the recipient protein, generating the phosphoprotein, ADP or AMP, plus an inorganic phosphate as reaction products. The amino acids phosphorylated in mammalian systems are referred to as O-phosphates and include tyrosine, threonine and serine [100]. O-phosphates are phosphoproteins in which the hydrogen on the hydroxyl moiety of the R-group has been substituted with an anionic phosphate group. More than one of these phosphoaminoacids within a given protein may be phosphorylated, as is the case with activated MAP kinases, which are dually phosphorylated on both threonine and tyrosine residues [101]. While the three O-phosphates are, by far, the most commonly studied, other phosphoaminoacids, such as phosphohistidine, also exist [100]. Phosphoaminoacids that flank the nuclear localization signal within a protein can affect the affinity of a transport receptor for the NLS on phosphorylation. For example, phosphorylation of the residues adjacent to the NLS of the SV40 large T antigen regulates nuclear import of the large T antigen [102].
The removal of phosphate groups is performed by protein phosphatases [103]. Phosphatases can be regulated by calcium and calmodulin [104] and can be classified into four different families based on substrate specificity and degree of conservation within their catalytic domains [105]. With respect to nuclear transport, dephosphorylation functions in a manner similar to that of phosphorylation. In this case, the removal of a phosphate group can unmask the transport motif, leading to its recognition and binding by the appropriate transport receptor. For example, dephosphorylation of NFAT exposes its NLS, subsequently leading to its nuclear translocation [87]. Furthermore, recent work has identified an association between protein phosphatase 2A and members of the importin-β superfamily [106], which may indicate the existence of generalized phosphatase-dependent regulation on nuclear transport machinery.
Regulation of nucleocytoplasmic transport occurs within the cytosol and possibly at the NE. Both kinases and phosphatases have been identified to reside within the nuclear pore [107]. A brief overview of transport receptors, their subclasses, and a short review on the NPC are presented in the following sections.
Nuclear Transport Receptors
The proteins responsible for the movement of molecules back and forth between the nuclear and cytosolic compartments are collectively referred to as importins [108]. Identification of the first importins, also known as karyopherins, occurred in the early 1990s. It was the isolation and characterization of importin-β (or karyopherin β1) [109] that unequivocally demonstrated the requirement of a cytosolic factor in the nuclear import process. As more transport receptors were discovered in subsequent years, it became apparent that all of them could be categorized into one family. The importin-β superfamily is the de facto designation describing these related proteins. For mechanistic reasons, transport receptors responsible for the export of molecules out of the nucleus into the cytoplasm also fall within this importin superfamily [108].
Importin-β
The prototypical importin-β protein was first isolated and identified as a 97kDa molecule in bovine erythrocytes [109], and was subsequently found to be essential to nuclear protein import [109,110]. Structural analyses of importin-β revealed that it contains 19 tandem HEAT repeats, arranged so that the tertiary conformation of the molecule is a right-handed superhelical protein with a high degree of flexibility [111]. Functionally, it can bind to nuclear pore proteins containing the FxFG motif and possesses an amino-terminal Ran binding domain [112-114]. With respect to nuclear import, importin-β participates as part of a trimeric complex formed when a protein bearing a ‘classical’ polybasic NLS is recognized by its receptor, importin-α [115]. Interaction of the NLS with its cognate receptor causes importin-α to undergo a conformational shift which then promotes its binding to the carboxy terminus of importin-β by virtue of an importin-β binding (IBB) domain at its N-terminus [111,116]. Formation of the importin heterodimer causes mutual conformational changes in both subunits which ultimately promote nuclear protein import [116] in a Ran and energy dependent manner.
Alternatively, a protein may directly bind importin-β without the use or need of adaptor proteins such as importin-α [108]. Several viral proteins [117-119] are able to bind directly to the IBB domain and form a heterodimeric complex which is imported into the nucleus. Additionally, other endogenous eukaryotic proteins use this mechanism to be imported [120,121]. Based on these observations, the importin-β superfamily can be subdivided into two categories: those that are capable of binding a molecule directly for import/export; and those that utilize adaptor proteins. Adaptor proteins themselves can be further classified into importin-α type NLS receptors, or nonimportin- α type receptors.
Adaptors: importin-α
Importin-α is the prototypical adaptor protein that recognizes both mono- and bipartite ‘classical’ NLSs [122,123] and was simultaneously isolated and identified with importin-β, as mentioned previously (109). This molecule consists of 10 tandem ARM motifs, with the main NLS recognition sites residing within the second and fourth ARM repeats [124]. At the C-terminus is an acidic domain required for binding to CAS, the endogenous nuclear exporter of importin-α [125-127]. At its N-terminus, importin-α possesses the IBB domain required for interaction with importin-β [111,127].
Identification of a polybasic sequence within the IBB [128] led to the hypothesis that importin-α may also possess autoinhibitory activity in addition to binding to importin-β. This was confirmed by a later study which reported that a Lys-Arg-Arg sequence within the IBB domain binds to the NLS recognition domain within importin-α [129]. This highly conserved sequence was identified in several forms of importin-α from yeast, mouse and human sources [129]. Using a variety of plasmids encoding different mutations of this IBB domain, Harreman et al [129] demonstrated that this sequence was essential in promoting NLS-cargo release within the nucleus and it was suggested that the autoinhibitory properties of the IBB domain prevents futile cycling of unloaded importin-α/β complexes.
More than one form of importin-α exists. In humans, six forms have been identified, each of which are separate gene products [130], as is the case for the various mouse isoforms of importin-α that have been identified [131]. Since tissue specific distribution of the importin-α isoforms has been observed, it is possible that each isoform is responsible for the import of a specific group or subset of proteins. For example, RanGEF/RCC1 is a constitutively nuclear enzyme that is preferentially imported by importin-α3 [130,132]. Future studies investigating specific importins and their respective cargoes will provide a more comprehensive understanding of the diversity of import receptors involved in nuclear transport.
Adaptors: non-importin-α receptors
There are other adaptor proteins with specific cargoes distinct from those mediated by the importin α/β complex. In some ways, these molecules possess similar characteristics to those of importin-α. For example, they possess an IBB domain as well as the ability to bind to their respective cargoes. However, the recognition motif by which they bind is typically quite different.
Snurportin1 was identified as the adaptor responsible for importing m3G capped U snRNPs into nuclei [133]. Similar to importin-α, snurportin1 possesses an IBB domain. However, it is distinct from the former in that it allows Ran- and energy- independent nuclear protein import [134]. In order to illustrate that this ability was characteristic of the snurportin1 IBB, Huber et al [133] generated a snurportin1 mutant that possessed the importin-α IBB and demonstrated that ribonucleoprotein import became Ran dependent.
Histone H1 is a small (~22 kDa) basic protein that is consitutively nuclear and actively transported despite its small size [135-137]. Also known as linker histones, they are involved in both the maintenance of chromatin structure as well as transcriptional regulation [137] and are the exclusive ligands of the importin-7 receptor [136,138]. Several portions of histone H1 possess stretches of basic amino acids that, on their own, serve as NLSs that can functionally interact with individual import factors (136). However, import of the entire complex requires a heterodimeric receptor composed of both importin-β and importin-7, the latter being the adaptor specific for histone H1 [136]. It is believed that in addition to serving as an import receptor, the importin-β/-7 complex may also serve as a chaperone for histone H1 [136].
The XRIPα adaptor protein is responsible for importing replication protein A (RPA) [139]. RPA is a single stranded DNA binding protein composed of three subunits (140) that is essential for chromosomal DNA replication [141], repair [142] and recombination [143-145]. The adaptor was originally identified using a yeast two hybrid assay by Jullien et al [139]. Briefly, several domains of the RPA holoenzyme were fused to the Gal4 DNA-binding domain to form the “bait” constructs that were used to screen a Xenopus oocyte cDNA library. The identified adaptor was found to specifically bind RPA and possessed an IBB structurally distinct from that of the importin-α IBB [139]. Similar to previously characterized importin-β binding domains, the IBB of XRIPα is located at the N-terminus and contains an arginine-rich, basic motif [139]. The similarities end there, however, as arginines in this particular receptor do not form continuous stretches of basic amino acids as found in the IBBs of HIV Tat and Rev proteins [119].
It is clear from these examples that although members within the importin family share some properties, they possess individualized characteristics which contribute to the recognition of a broad and diverse array of substrates. While this observation has been made for molecules that mediate nuclear import, it also holds true for the members of the importin-β superfamily that are responsible for nuclear export.
Exportins
Exportins are members of the importin-β family that are responsible for the movement of proteins, RNA and ribonucleoproteins from the nucleus into the cytoplasm [107,146]. They are generally similar to the importins in that they recognize a localization signal, bind to Ran and interact with proteins of the NPC. On closer investigation of the exportin molecule, differences become apparent.
CRM1/exportin-1 is the prototypical exportin molecule responsible for nuclear transport of the bulk of exported substrates. It recognizes a nuclear export signal (NES), which is different from the polybasic NLS mentioned earlier [147]. NESs are typically leucine rich sequences found on a protein destined for transport into the cytosol [148]. The first such NES was identified in the HIV Rev protein (149). In nuclear export, CRM1 binds to the NES and associates with RanGTP to form a stable export complex, similar to the mechanism of nuclear import. This heterotrimeric complex is then directed to the nuclear basket on the nucleoplasmic face of the NPC [76] and subsequently exported to the cytoplasm. Once there, RanGAP activates the intrinsic GTPase activity of Ran [150]. Hydrolysis of GTP to form RanGDP promotes dissociation of the nuclear export complex, releasing the cargo into the cytoplasm. Although this pathway is recognized as the ‘workhorse’ pathway of nuclear export, other CRM1- independent transport mechanisms exist.
One such example is the nuclear export of importin-α. Kutay et al [126] identified CAS as an essential protein required to transport importin-α from the nucleus back to the cytosol following a round of nuclear import. Subsequent studies confirmed the functionality of CAS as a nuclear exporter [151-153]. CAS activity can be modulated by sphingolipids like ceramide to alter nuclear protein import [154]. Calreticulin is another nuclear export receptor identified by Holaska et al [155]. Although indirectly involved in nuclear import of NFAT3 [156] and MEF2C [157], calreticulin directly exports nuclear, steroid, non-steroid hormone and orphan receptors [158,159] from the nucleus in a calcium-dependent manner [160]. This observation was surprising, given the constitutive localization of calreticulin to the endoplasmic/sarcoplasmic reticulum [161]. Calreticulin possesses an endoplasmic reticulum (ER)/sarcoplasmic reticulum retention signal which limits its presence in the cytosol [161]. Walther et al [158] examined calreticulin-mediated export of the glucocorticoid receptor (GR) using a heterokaryon assay and reported that transient permeabilization of the ER membrane allowed calreticulin to leave the ER lumen. Calreticulin release led to increased nuclear export of GR compared with rates observed in control cells and it was hypothesized that calreticulin-mediated export was a normal physiological mechanism used to rapidly clear the nucleus of receptors before mitosis [158]. The role of calreticulin as a nuclear export receptor remains debatable because no identifiable nuclear localization signal or import mechanism has been characterized for calreticulin.
Proteins are not the only molecules subject to exportin mediated transport. The export of RNA species involves completely different sets of receptors. The transport of transfer RNAs (tRNAs) is mediated specifically by exportin-t [162,163] and to a lesser extent, exportin-5 [164]. Both exportin-t and exportin-5 cooperatively bind RanGTP and the tRNA molecule directly, without the need for adaptors. Similar to tRNAs, mRNAs possess specific proteins which direct them out of the nucleus in a CRM1- and Ran- independent manner.
Messenger RNA is delivered to the cytosol by the TAP receptor [165]. This exportin forms a complex with another molecule, NXT1, to form a heterodimer that mediates the movement of mRNA out of the nucleus [166-168]. However, transport of mRNA is slightly more complex, as other factors which participate in splicing, elongation and termination of the nascent mRNA transcript are required to mediate the association between the TAP/NXT1 complex and mRNA [146,169,170].
The diverse nature of nuclear transport receptors and adaptors justifies the premise of the existence of other receptors that remain to be identified [171]. Also, while knowledge concerning nucleocytoplasmic trafficking control has come primarily from studies that have examined regulatory processes on the cytosolic machinery, evidence suggests that modifications at the membrane bound NPC also regulate nuclear trafficking [172].
Ran and Nuclear Transport
Ran is a small GTPase intimately involved with nucleocytoplasmic trafficking (Figure 2) [70,142,173,174]. Located predominantly in the nucleus, it is capable of shuttling back and forth between the nuclear and cytoplasmic compartments [175,176]. Nuclear Ran is bound to GTP and delivered to the cytosol in conjunction with an export complex [177]. Once there, it is converted to RanGDP [150,178]. This is extremely important, as hydrolysis of RanGTP promotes the dissociation of exportins from their cargo when they reach the cytosol.
Figure 2: Ran cycle. The classic mechanism of import requires a functional Ran cycle. The Ran gradient across the nuclear envelope provides directionality and motive force to drive nuclear transport. RanGDP is imported into the nucleus by NTF2, where GDP is exchanged for GTP by RanGEF/RCC1 (see text). RanGTP is then exported from the nucleus, either in conjunction with an export complex or with importin-β. In the cytosol, RanGAP coactivates Ran, generating RanGDP and the cycle continues. CAS is the import receptor for importin-β and recycles it back to the cytosol.
While cytoplasmic delivery of Ran occurs via its participation in an export complex, import of Ran requires the RanGDP specific import factor called p10/NTF2. First identified in S. cerevisiae by Nehrbass and Blobel [179], this molecule was shown by these and other authors to bind to the FxFG repeats of isolated nups (180,181). Further work clarified the role of p10/NTF2 as a nuclear import receptor for Ran in its GDP bound form [182-184], as it was observed that NTF2 specifically bound RanGDP and not RanGTP [179]. NTF2 is located primarily at the NE, bound to nups [181], since the Ran-binding and nup binding domains are discrete regions of the protein (180). It is also distributed between the cytoplasm and the nucleus [180]. The concentration of NTF2 at the NE is on the order of ~20 μM and it exists in a dimerized form, whereas it is monomeric in the cytoplasm and the nucleus, at a concentration of 0.3 μM and 0.6 μM, respectively [180]. These differences in concentration affect the monomer-dimer equilibrium of NTF2, as Chaillan- Huntington et al reported a dissociation constant of ~1.1 μM for NTF2 [180]. At concentrations below this KD, the dimer dissociates and significantly weakens the affinity of NTF2 for RanGDP [180], releasing it into the nucleoplasm, which can then be recharged with GTP by RCC1.
Ran has weak inherent GDP/GTP exchange activity as well as intrinisic GTP hydrolyzing capabilities [185]. In order for Ran to maintain its distinct GTP and GDP bound states in the nucleus and cytoplasm, respectively, a host of accessory proteins exist which regulate and enhance both the hydrolyzing ability of Ran as well as its nucleoside binding capacity. Within the nucleus is the RanGEF/RCC1 enzyme. It is constitutively imported and maintains its nuclear localization by associating with chromatin (186). This enzyme is responsible for replacing GDP with GTP, and maintains Ran in a GTP bound form [187]. Binding of RanGTP to importin-β entering the nucleus promotes dissociation of the import complex and subsequent release of the NLS bearing cargo into the nucleus [188]. Export complexes are formed only in the presence of RanGTP [177,189]. RCC1 is imported into the nucleus by two distinct mechanisms – one which depends on importin-α3 [130] and another that does not depend on energy or preexisting Ran gradients [190].
RanGTP that exits the nucleus in association with an export complex is converted to RanGDP by the concerted action of several coactivating proteins. Chief among these proteins is RanGAP1 [150], which, together with the Ran binding protein RanBP1 [191], increases the intrinsic hydrolytic activity of Ran. RanGAP1 exists in two forms, a soluble cytosolic form and a SUMOylated, NPCassociated form [91,92]. At the NPC, another Ran binding protein called RanBP2/Nup358 is part of the cytoplasmic filaments of the complex which also acts co-operatively with SUMO-RanGAP1 [91,92]. These elements maintain Ran in a GDP bound form in the cytoplasm. The nuclear components of the Ran cycle maintain high nuclear concentrations of RanGTP. It has been estimated that the concentration of RanGTP is over 200-fold greater in the nucleus than in the cytoplasm [192]. Together, the cytoplasmic and nuclear elements of the Ran cycle establish a steep RanGTP gradient across the NE. This is the source of energy for active nuclear transport as well as the element that specifies the vectorial movement of transport cargo.
The majority of nucleocytoplasmic trafficking requires a functional Ran cycle to operate (Figure 2). Exceptions exist, however, and Ran independent transport has been identified for a variety of different proteins [193,194]. The nuclear import receptor transportin enters the nucleus by a Ran unassisted mechanism [194]. IκBα, a regulator of NFκB transport, is imported without Ran [195]. Other non-transport proteins, such as beta catenin [196] and the U1A and U2B'' spliceosome proteins [197] also enter the nucleus by a pathway independent of Ran. RanGAP activity can be stimulated by lipid molecules like lysophosphatidylcholine [198].
The Future of Research on Nucleocytoplasmic Trafficking
Transport of proteins in and out of the nucleus is rapidly becoming recognized as a key process in the life and death of a cell [199]. It is also a critical pathway in the capacity of a cell to adapt to an everchanging environment [199]. With such a central role in the viability and function of a cell, it is not surprising that scientists are becoming increasingly aware of its potential role in cell pathology [200-203] and its identifications as a target for therapeutic interactions [204]. It is predicted that nucleocytoplasmic trafficking will become an increasingly important and attractive subject in cell biology.
Disclosures
The authors have no financial disclosures or conflicts of interest to declare.
References
- Pante N, Aebi U. Molecular dissection of the nuclear pore complex. Crit Rev Biochem Mol Biol 1996;31:153-99.
- Forbes DJ. Structure and function of the nuclear pore complex. Annu Rev Cell Biol 1992;8:495-527.
- Hinshaw JE. Architecture of the nuclear pore complex and its involvement in nucleocytoplasmic transport. Biochem Pharmacol 1994;47:15-20.
- Hinshaw JE, Carragher BO, Milligan RA. Architecture and design of the nuclear pore complex. Cell 1992;69:1133-41.
- Panté N, Aebi U. The nuclear pore complex. J Cell Biol 1993;122:977-84.
- Panté N, Aebi U. Exploring nuclear pore complex structure and function in molecular detail. J Cell Sci 1995;19(Suppl):1-11.
- Adam SA. The nuclear pore complex. Genome Biol 2001;2:1-6.
- Miller M, Park MK, Hanover JA. Nuclear pore complex: Structure, function, and regulation. Physiol Rev 1991;71:909-49.
- Ryan KJ, Wente SR. The nuclear pore complex: A protein machine bridging the nucleus and cytoplasm. Curr Opin Cell Biol 2000;12:361-71.
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J Cell Biol 2000;148:635-51.
- Panté N, Thomas F, Aebi U, Burke B, Bastos R. Recombinant Nup153 incorporates in vivo into Xenopus oocyte nuclear pore complexes. J Struct Biol 2000;129:306-12.
- Akey CW, Radermacher M. Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J Cell Biol 1993;122:1-19.
- Danker T, Oberleithner H. Nuclear pore function viewed with atomic force microscopy. Pflugers Arch 2000;439:671-81.
- Panté N, Kann M. Nuclear pore complex is able to transport macromolecules with diameters of ~39 nm. Mol Biol Cell 2002;13:425-34.
- Feldherr C, Akin D, Moore MS. The nuclear import factor p10 regulates the functional size of the nuclear pore complex during oogenesis. J Cell Sci 1998;111:1889-96.
- Feldherr C , Akin D, Cohen RJ. Regulation of functional nuclear pore size in fibroblasts. J Cell Sci 2001;114:4621-7.
- Perez-Terzic C, Jaconi M, Clapham DE. Nuclear calcium and the regulation of the nuclear pore complex. Bioessays 1997;19:787-92.
- Perez-Terzic C, Pyle J, Jaconi M, Stehno-Bittel L, Clapham DE. Conformational states of the nuclear pore complex induced by depletion of nuclear Ca2+stores. Science 1996;273:1875-7.
- Mazzanti M, Bustamante JO, Oberleithner H. Electrical dimension of the nuclear envelope. Physiol Rev 2001;1:1-19.
- Keminer O, Peters R. Permeability of single nuclear pores. Biophys J 1999;77:217-28.
- Shahin V, Danker T, Enss K, Ossig R, Oberleithner H. Evidence for Ca2+- and ATP-sensitive peripheral channels in nuclear pore complexes. FASEB J 2001;15:1895-901.
- Stoffler D, Feja B, Fahrenkrog B, Walz J, Typke D, Aebi U. Cryo-electron tomography provides novel insights into nuclear pore architecture: Implications for nucleocytoplasmic transport. J Mol Biol 2003;328:119-30.
- Sharma A, Solmaz S R, Blobel G, Melcak I. Ordered regions of nucleoporins Nup62, Nup54, and Nup58 form dynamic complexes in solution. J. Biol Chem 2015; 290:18370-8.
- Solmaz S R, Blobel G, Melcak I. Ring cycle for dilating and constricting the nuclear pore. Proc Natl Acad Sci USA 2013;110:5858-63.
- Yokoyama N, Hayashi N, Seki T, et al. A giant nucleopore protein that binds Ran/TC4. Nature 1995;376:184-8.
- Yaseen N R, Blobel G. Two distinct classes of Ran-binding sites on the nucleoporin Nup-358. Proc Natl Acad Sci USA 1999;96:5516-21.
- Yaseen N R, Blobel G. GTP hydrolysis links initiation and termination of nuclear import on the nucleoporin Nup358. J Biol Chem 1999;274:26493-26502.
- Wu J, Matunis M J, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran- GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine rich region. J Biol Chem 1995;270:14209-13.
- Goldberg M W, Allen T D. High resolution scanning electron microscopy of the nuclear envelope: Demonstration of a new, regular, fibrous lattice attached to the baskets of the nucleoplasmic face of the nuclear pores. J Cell Biol 1992;119:1429-40.
- Fahrenkrog B, Maco B, Fager AM, et al. Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J Struct Biol 2002:140:254-67.
- Daigle N, Beaudouin J, Hartnell L, et al. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J Cell Biol 2001;154:71-84.
- Moore-Nichols D, Arnott A, Dunn RC. Regulation of nuclear pore complex conformation by IP3 receptor activation. Biophys J 2002;83:1421-8.
- Perez-Terzic C, Gacy A M, Bortolon R, et al. Structural plasticity of the cardiac nuclear pore complex in response to regulators of nuclear import. Circ Res 1999;84:1292-301.
- Goldberg MW, Rutherford SA, Hughes M, et al. Ran alters nuclear pore complex conformation. J Mol Biol 2000;300:519-29.
- Stoffler D, Goldie K N, Feja B, Aebi U. Calcium-mediated structural changes of native nuclear pore complexes monitored by time-lapse atomic force microscopy. J Mol Biol 1999;287:741-52.
- Pascual-Garcia P, Capelson M. Nuclear pores as versatile platforms for gene regulation. Curr Opin Genet Dev 2014;25:110-7.
- Capelson M, Hetzer M W. The role of nuclear pores in gene regulation, development and disease. EMBO Rep 2009;10:697-705.
- Bustamante JO, Michelette ER, Geibel JP, Dean DA, Hanover JA, McDonnell TJ. Calcium, ATP and nuclear pore channel gating. Pflügers Arch 2000;439:433-44.
- Walther T C, Askjaer P, Gentzel M, et al. RanGTP mediates nuclear pore complex assembly. Nature. 2003;424:689-94.
- Harel A, Chan R C, Lachish-Zalait A, Zimmerman E, Elbaum M, Forbes D J. Importin β negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol Biol Cell 2003;14:4387-96.
- Cordes VC, Rackwitz HR, Reidenbach S. Mediators of nuclear protein import target karyophilic proteins to pore complexes of cytoplasmic annulate lamellae. Exp Cell Res 1997;237:419-33.
- Imreh G, Hallberg E. An integral membrane protein from the nuclear pore complex is also present in the annulate lamellae: implications for annulate lamellae formation. Exp Cell Res 2000;259:180-90.
- Ewald A, Kossner U, Scheer U, Dabauvalle MC. A biochemical and immunological comparison of nuclear and cytoplasmic pore complexes. J Cell Sci 1996;109:1813-24.
- Belgareh N, Doye V. Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells. J Cell Biol 1997;136:747-59.
- Toyama BH, Savas JN, Park SK, et al. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 2013;154:971-82.
- Favreau C, Bastos R, Cartaud J, Courvalin J C, Mustonen P. Biochemical characterization of nuclear pore complex protein gp210. Eur J Biochem. 2001;268:3883-9.
- Kihlmark M, Imreh G, Hallberg E. Sequential degradation of proteins from the nuclear envelope during apoptosis. J Cell Sci 2001;114:3643-3653.
- Lenz-Böhme B, Wismar J, Fuchs S, et al. Insertional mutation of the Drosophila nuclear lamin Dm0 gene results in defective nuclear envelopes, clustering of nuclear pore complexes, and accumulation of annulate lamellae. J Cell Biol 1997;137:1001-16.
- Heath CV, Copeland CS, Amberg DC, Priore VD, Snyder M, Cole CN. Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutation of the Saccharomyces cerevisiae RAT2/NUP120 gene. J Cell Biol 1995;131:1677-97.
- Aitchison JD, Blobel G, Rout MP. Nup120p: A yeast nucleoporin required for NPC distribution and mRNA transport. J Cell Biol 1995;131:1659-75.
- Pemberton L, Rout MP, Blobel G. Disruption of the nucleoporin gene NUP133 results in clustering of nuclear pore complexes. Proc Natl Acad Sci USA 1995;92:1187-91.
- Maul GD, Deaven L. Quantitative determination of nuclear pore complexes in cycling cells with differing DNA content. J Cell Biol 1977;73:748-60.
- Maul GG, Deaven LL, Freed JJ, Campbell GL, Beçak W. Investigation of the determinants of nuclear pore number. Cytogenet Cell Genet 1980;26:175-90.
- Maul HM, Hsu BY, Borun TM, Maul GG. Effect of metabolic inhibitors on nuclear pore formation during the HeLa S3 cycle. J Cell Biol 1973;59:669-76.
- Iouk T, Kerscher O, Scott R J, Basrai MA, Wozniak RW. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J Cell Biol 2002;159:807-19.
- Salina D, Enarson P, Rattner J B, Burke B. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J Cell Biol 2003;162:991-1001.
- Stukenberg P T, Macara I G. The kinetochore NUPtials. Nat Cell Biol 2003;5:945-7.
- Rout MP, Wente SR. Pores for thought: Nuclear pore complex proteins. Trends Cell Biol 1994;4:357-65.
- Wu X, Kasper LH, Mantcheva RT, Mantchev GT, Springett MJ, vanDeursen JM. Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function. Proc Natl Acad Sci USA 2001;98:3191-6.
- Powers MA, Forbes DJ, Dahlberg JE, Lund E. The vertebrate GLFG nucleoporin, Nup98, is an essential component of multiple RNA export pathways. J Cell Biol 1997;136:241-50.
- Bayliss R, Littlewood T, Stewart M. Structural basis for the interaction between FxFG nucleoporin repeats and importin-β in nuclear trafficking. Cell 2000;102:99-108.
- Ando D, Zandi R, Kim Y W, Colvin M, Rexach M, Gopinathan A. Nuclear pore complex protein sequences determine overall copolymer brush structure and function. Biophys J 2014;1997-2007.
- Lim RY, Fahrenkrog B, Koser J, Schwarz-Herion K, Deng J, Aebi U. Nanomechanical basis of selective gating by the nuclear pore complex. Science 2007;318:640-3.
- Ribbeck K, Görlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J 2001;20:1320-30.
- Frey S, Richter RP, Gorlich D. FS-rich repeats of nuclear pore proteins from a three-dimensional mesh work with hydrogel-like properties. Science 2006;314:815-7.
- Yamada J, Phillips JL, Patel S, et al. A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol Cell Proteomics 2010;9:2205-24.
- Izaurralde E, Kutay U, vonKobbe C, Mattaj I W, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J 1997;16:6535-47.
- Pu RT, Dasso M. The balance of RanBP1 and RCC1 is critical for nuclear assembly and nuclear transport. Mol Biol Cell 1997;8:1955-70.
- Avis JM, Clarke PR. Ran, a GTPase involved in nuclear processes: Its regulators and effectors. J Cell Sci 1996;109:2423-7.
- Azuma Y, Dasso M. The role of Ran in nuclear function. Curr Opin Cell Biol 2000;12:302-7.
- Nachury MV, Weis K. The direction of transport through the nuclear pore can be inverted. Proc Natl Acad Sci USA 1999;96:9622-7.
- Ben-Efraim I, Gerace L. Gradient of increasing affinity of importin beta for nucleoporins along the pathway of nuclear import. J Cell Biol 2001;152:411-7.
- Kehlenbach R H, Dickmanns A, Kehlenbach A, Guan T, Gerace L. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J Cell Biol 1999;145:645-57.
- Singh BB, Patel HH, Roepman R, Schick D, Ferreira P A. The zinc finger cluster domain of RanBP2 is a specific docking site for the nuclear export factor, exportin-1. J Biol Chem 1999;274:37370-37378.
- Shah S, Forbes DJ. Separate nuclear import pathways converge on the nucleoporin Nup153 and can be dissected with dominant- negative inhibitors. Curr Biol 1998;8:1376-86.
- Ullman KS, Shah S, Powers MA, Forbes DJ. The nucleoporin nup153 plays a critical role in multiple types of nuclear export. Mol Biol Cell 1999;10:649-64.
- Kapinos LE, Schoch RL, Wagner RS, Schleicher KD, Lim RY. Karyopherin-centric control of nuclear pores based on molecular occupancy and kinetic analysis of multivalent binding with FG nucleoporins. Biophys J 2014;106:1751-62.
- Schoch RL, Kapinos LE, Lim RY. Nuclear transport receptor binding avidity triggers a self-healing collapse transition in FG-nucleoporin molecular brushes. Proc Natl Acad Sci USA 2012;109:16911-6.
- Zwerger M, Eibauer M, Medalia O. Insights into the gate and scaffold of the nuclear pore complex. Nucleus 2016;7:1-7.
- Tu LC, Musser SM. Single molecule studies of nucleocytoplasmic transport. Biochem Biophys Acta 20111813:1607-18.
- Kalderon D, Richardson WD, Markham AF, Smith A E. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 1984;311:33-8.
- Dingwall C, Sharnick SV, Laskey RA. A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell 1984;30:449-58.
- Dingwall C, Dilworth SM, Black SJ, Kearsey SE, Cox LS, Laskey RA. Nucleoplasmin cDNA sequence reveals polyglutamic acid tracts and a cluster of sequences homologous to putative nuclear localization signals. EMBO J 1987;6:69-74.
- Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol 1995;129:551-60.
- Henderson B R, Eleftheriou A. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp Cell Res 2000;256:213-224.
- Cullen BR. Nuclear RNA export. J Cell Sci 2003;116:587-97.
- Zhu J, Shibasaki F, Price R, et al. Intramolecular masking of nuclear import signal on NF-AT4 by casein kinase I and MEKK1. Cell 1998;93:851-61.
- Okamura H, Aramburu J, Garcia-Rodriguez C, et al. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol Cell 2000;6:539-50.
- Hay RT, Vuillard L, Desterro JM, Rodriguez MS. Control of NF-kappa B transcriptional activation by signal induced proteolysis of I kappa B alpha. Philos Trans R Soc Lond B Biol Sci 1999;354:1601-9.
- Rihs HP, Jans DA, Fan H, Peters R. The rate of nuclear cytoplasmic protein transport is determined by the casein kinase II site flanking the nuclear localization sequence of the SV40 T-antigen. EMBO J 1991;10:633-9.
- Matunis MJ, Wu J, Blobel G. SUMO-1 modification and its role in targeting the RanGTPase-activating protein, RanGAP1, to the nuclear pore complex. J Cell Biol 1998;140:499-509.
- Melchior F, Schergaut M, Pichler A. SUMO: Ligases, isopeptidases and nuclear pores. Trends Biochem Sci 2003;28:612-8.
- Tasken K, Aandahl EM. Localized effects of cAMP mediated by distinct routes of protein kinase A. Physiol Rev 2004;84:137-67.
- Feil R, Lohmann S M, deJonge H, Walter U, Hofmann F. Cyclic GMP-dependent protein kinases and the cardiovascular system: insights from genetically modified mice. Circ Res 2003;93:907-16.
- Schlossmann J, Feil R, Hofmann F. Signaling through NO and cGMP-dependent protein kinases. Ann Med 2003;35:21-7.
- Heist E K, Schulman H. The role of Ca2+/calmodulin-dependent protein kinases within the nucleus. Cell Calcium. 1998;23:103-14.
- Sugiura M, Kono K, Liu H, Shimizugawa T, Minekura H, Spiegel S, Kohama T. Ceramide kinase, a novel lipid kinase. Molecular cloning and functional characterization. J Biol Chem 2002;277:23294-300.
- Kanoh H ,Yamada K, Sakane F. Diacylglycerol kinases: emerging downstream regulators in cell signaling systems. J Biochem (Tokyo) 2002;131:629-33.
- Chahine MN, Blackwood DP, Dibrov E, et al. Oxidized low density lipoprotein affects smooth muscle cell growth through MAPK- medicated actions on nuclear protein import. J Mol Cell Cardiol 2009;46:431-41.
- Klumpp S, Krieglstein J. Phosphorylation and dephosphorylation of histidine residues in proteins. Eur J Biochem 2002;269:1067-71.
- Khokhlatchev AV, Canagarajah B, Wilsbacher J, et al. Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell 1998;93:605-15.
- Xiao C.-Y, Hübner S, Jans D A. SV40 large tumor antigen nuclear import is regulated by the double-stranded DNA-dependent protein kinase site (serine 120) flanking the nuclear localization sequence. J Biol Chem 1997;272:22191-8.
- Barford D, Das AK, Egloff MP. The structure and mechanism of protein phosphatases: Insights into catalysis and regulation. Annu Rev Biophys Biomol Struct 1998;27:133-64.
- Tonks N K, Cohen P. Calcineurin is a calcium ion-dependent, calmodulin-stimulated protein phosphatase. Biochim Biophys Acta 1983;747:191-3.
- Bollen M, Beullens M. Signaling by protein phosphatases in the nucleus. Trends Cell Biol 2002;12:138-45.
- Lubert EJ, Sarge KD. Interaction between protein phosphatase 2A and members of the importin β superfamily. Biochem Biophys Res Comm 2003;303:908-13.
- Faustino RS, Maddaford, TG, Pierce GN. Mitogen activated protein kinase at the nuclear pore complex. J Cell Mol Med 2011;15:928-37.
- Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607-60.
- Adam EJ, Adam SA. Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J Cell Biol 1994;125:547-55.
- Chi NC, Adam EJ, Adam SA. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J Cell Biol 1995;130:265-74.
- Cingolani G, Petosa C, Weis K, Müller C W. Structure of importin-β bound to the IBB domain of importin-α. Nature 1999;399:221-9.
- Kutay U, Izaurralde E, Bischoff FR, Mattaj I W, Görlich D. Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex. EMBO J 1997;16:1153-63.
- Chi NC, Adam SA. Functional domains in nuclear import factor p97 for binding the nuclear localization sequence receptor and the nuclear pore. Mol Biol Cell 1997;8:945-56.
- Chi NC, Adam EJ, Adam SA. Different binding domains for Ran- GTP and Ran-GDP/RanBP1 on nuclear import factor p97. J Biol Chem 1997;272:6818-22.
- Adam SA, Gerace L. Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell 1991;66:837-47.
- Cingolani G, Lashuel HA, Gerace L, Müller CW. Nuclear import factors importin α and importin β undergo mutually induced conformational changes upon association. FEBS Lett 2000;484:291-8.
- Palmeri D, Malim MH. Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol Cell Biol 1999;19.
- Henderson BR, Percipalle P. Interactions between HIV Rev and nuclear import and export factors: The Rev nuclear localisation signal mediates specific binding to human importin-beta. J Mol Biol 1997;274.
- Truant R, Cullen BR. The arginine-rich domains present in human immunodeficiency virus type 1 tat and rev function as direct importin beta-dependent nuclear localization signals. Mol Cell Biol 1999;19.
- Jäkel S, Görlich D. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J 1998;17:4491-502.
- Lam MH, Briggs LJ, Hu W, Martin TJ, Gillespie M T, Jans D A. Importin β recognizes parathyroid hormone-related protein with high affinity and mediates its nuclear import in the absence of importin α. J Biol Chem 1999;274:7391-8.
- Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell 1998;94:193-204.
- Fontes M R M, Teh T, Kobe B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α. J Mol Biol 2000;297:1173-94.
- Melén K, Fagerlund R, Franke J, Köhler M, Kinnunen L, Julkunnen I. Importin α nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J Biol Chem 2003;278:28193-200.
- Hood J K, Silver P A. Cse1p is required for export of Srp1p/ importin-α from the nucleus in Saccharomyces cerevisiae. J Biol Chem 1998;273:35142-6.
- Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell 1997;90:1061-71.
- Herold A, Truant R, Wiegand H, Cullen BR. Determination of the functional domain organization of the importin α nuclear import factor. J Cell Biol 1998;143:309-18.
- Kobe B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin α. Nat Struct Biol 1999;6:388-97.
- Harreman MT, Hodel MR, Fanara P, Hodel AE, Corbett AH. The auto-inhibitory function of importin α is essential in vivo. J Biol Chem. 2003;278:5854-5863.
- Köhler M, Speck C, Christiansen M, et al. Evidence for distinct substrate specificities of importin α family members in nuclear protein import. Mol Cell Biol 1999;19:7782-91.
- Kamei Y, Yuba S, Nakayama T, Yoneda Y. Three distinct classes of the α-subunit of the nuclear pore-targeting complex (importin-α) are differentially expressed in adult mouse tissues. J Histochem Cytochem 1999;47:363-72.
- Talcott B, Moore MS. The nuclear import of RCC1 requires a specific nuclear localization sequence receptor, karyopherin α3/Qip. J Biol Chem 2000;275:10099-104.
- Huber J, Cronshagen U, Kadokura M, et al. Snurportin1, an m3G- cap-specific nuclear import receptor with a novel domain structure. EMBO J 1998;17:4114-26.
- Huber J, Dickmanns A, Lührmann R. The importin-β binding domain of snurportin1 is responsible for the Ran- and energy- independent nuclear import of spliceosomal U snRNPs in vitro. J Cell Biol 2002;156:467-79.
- Belmont A. Dynamics of chromatin, proteins, and bodies within the cell nucleus. Curr Opin Cell Biol 2003;15:304-10.
- Bäuerle M, Doenecke D, Albig W. The requirement of H1 histones for a heterodimeric nuclear import receptor. J Biol Chem 2002;277:32480-9.
- Brown DT. Histone H1 and the dynamic regulation of chromatin function. Biochem Cell Biol 2003;81:221-7.
- Jäkel S, Albig W, Kutay U, et al. The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J 1999;18:2411-23.
- Jullien D, Görlich D, Laemmli UK, Adachi Y. Nuclear import of RPA in Xenopus egg extracts requires a novel protein XRIPα but not importin α. EMBO J 1999;18:4348-58.
- Wold MS. Replication protein A: Heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem 1997;66:61-92.
- dachi Y, Laemmli UK. Study of the cell cycle-dependent assembly of the DNA pre-replication centers in Xenopus egg extracts. EMBO J 1994;13:4153-64.
- Moore SP, Erdile L, Kelly T, Fishel R. The human homologous pairing protein HPP-1 is specifically stimulated by the cognate single-stranded binding protein hRP-A. Proc Natl Acad Sci USA 1991;88:9067-71.
- Shinohara A, Shinohara M, Ohta T, Matsuda S. Ogawa T. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells 1998;3:145-56.
- Longhese MP, Plevani P, Lucchini G. Replication factor A is required in vivo for DNA replication, repair and recombination. Mol Cell Biol 1994;14:7884-90.
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 1998;391:407-10.
- Fried H M, Kutay U. Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci 2003;60:1659-88.
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 1997;90:1051-60.
- Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell 1995;82:463-73.
- Fischer U, Huber J, Boelens WC, Mattaj I W, Lührmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 1995;82:475-83.
- Bischoff F R, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. RanGAP1 induces GTPase activity of nuclear ras-related Ran. Proc Natl Acad Sci USA 1994;91:2587-91.
- Scherf U, Kalab P, Dasso M, Pastan I, Brinkmann U. The hCSE1/ CAS protein is phosphorylated by HeLa extracts and MEK-1: MEK-1 phosphorylation may modulate the intracelluar localization of CAS. Biochem Biophys Res Comm 1998;250:623-8.
- Scherf U, Pastan I, Willingham M C, Brinkmann U. The human CAS protein which is homologous to the CSE1 yeast chromosome segregation gene product is associated with microtubules and mitotic spindle. Proc Natl Acad Sci USA 1996;93:2670-4.
- Ogryzko V V, Brinkmann E, Howard B H, Pastan I, Brinkmann U. Antisense inhibition of CAS, the human homologue of the yeast chromosome segregation gene CSE1, interferes with mitosis in HeLa cells. Biochemistry. 1997;36:9493-9500.
- Faustino RS, Cheung P, Richard MN, et al. Ceramide regulation of nuclear protein import. J Lipid Res 2008;49:654-662
- Holaska JM, Black BE , Love D C, Hanover JA, Leszyk J, Paschal BM. Calreticulin is a receptor for nuclear export. J Cell Biol 2001;152:127-40.
- Mesaeli N, Nakamura K, Zvaritch E, et al. Calreticulin is essential for cardiac development. J Cell Biol 1999;144:857-68.
- Li J, Pucéat M, Perez-Terzic C, et al. Calreticulin reveals a critical Ca2+ checkpoint in cardiac myofibrillogenesis. J Cell Biol 2002;158:103-13.
- Walther RE, Lamprecht C, Ridsdale A, et al. Nuclear export of the glucocorticoid receptor is accelerated by cell fusion-dependent release of calreticulin. J Biol Chem 2003;278:37858-64.
- DeFranco D B. Nuclear export: DNA-binding domains find a surprising partner. Curr Biol 2001;11:R1036-R1037.
- Holaska J M, Black B E, Rastinejad F, Paschal B M. Ca2+-dependent nuclear export mediated by calreticulin. Mol Cell Biol 2002;22:6286-97.
- Michalak M, Corbett E F, Mesaeli N, Nakamura K, Opas M. Calreticulin: One protein, one gene, many functions. Biochem J 1999;344:281-92.
- Arts GJ, Kuersten S, Romby P, Ehresmann B, Mattaj I W. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J 1998;17:7430-41.
- Kutay U, Lipowsky G, Izaurralde E, et al. Identification of a tRNA- specific nuclear export receptor. Mol Cell 1998;1:359-69.
- Calado A, Treichel N, Müller EC, Otto A, Kutay U. Exportin-5- mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J 2002;21:6216-24.
- Schmitt I, Gerace L. In vitro analysis of nuclear transport mediated by the C-terminal shuttle domain of Tap. J Biol Chem 2001;276.
- Lévesque L, Guzik B, Guan T, et al. RNA export mediated by Tap involves NXT1-dependent interactions with the nuclear pore complex. J Biol Chem 2001;276:44953-62.
- Guzik BW, Levesque L, Prasad S, et al. NXT1 (p15) is a crucial cellular cofactor in TAP-dependent export of intron-containing RNA in mammalian cells. Mol Cell Biol 2001;21:2545-54.
- Wiegand HL, Coburn GA, Zeng Y, Kang Y, Bogerd HP, Cullen BR. Formation of Tap/NXT1 heterodimers activates Tap-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes. Mol Cell Biol 2002;22:245-56.
- Gilbert W, Guthrie C. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol Cell 2004;13:201-212.
- Hurt E, Luo M J, Rother S, Reed R, Strasser K. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc Natl Acad Sci USA 2004;101:1858-62.
- Dean KA, vonAhsen O, Görlich D, Fried HM. Signal recognition particle protein 19 is imported into the nucleus by importin 8 (RanBP8) and transportin. J Cell Sci 2001;114:3479-85.
- Kehlenbach R H, Gerace L. Phosphorylation of the nuclear transport machinery down-regulates nuclear protein import in vitro. J Biol Chem. 2000;275:17848-56.
- Stochaj U, Rother KL. Nucleocytoplasmic trafficking of proteins: with or without Ran? Bioessays 1999;21:579-589.
- Melchior F, Paschal B, Evans J, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol 1993;123:1649-59.
- Quimby BB, Lamitina T, L’Hernault SW, Corbett AH. The mechanism of Ran import into the nucleus by nuclear transport factor 2. J Biol Chem 2000;275:28575-28582.
- Yoneda Y, Hieda M, Nagoshi E, Miyamoto Y. Nucleocytoplasmic protein transport and recycling of Ran. Cell Struct Funct 1999;24:425-33.
- Richards SA, Carey KL, Macara IG. Requirement of guanosine triphosphate-bound Ran for signal-mediated nuclear protein export. Science 1997;276:1842-4.
- Corbett AH, Koepp DM, Schlenstedt G, Lee MS, Hopper AK, Silver PA. Rna1p, a Ran/TC4 GTPase activating protein, is required for nuclear import. J Cell Biol 1995;130:1017-26.
- Nehrbass U, Blobel G. Role of the nuclear transport factor p10 in nuclear import. Science 1996;272:120-2.
- Chaillan-Huntington C, Braslavsky CV, Kuhlmann J, Stewart M. Dissecting the interactions between NTF2, RanGDP, and the nucleoporin XFXFG repeats. J Biol Chem 2000;275:5874-9.
- Bayliss R, Ribbeck K, Akin D, et al. Interaction between NTF2 and xFxFG-containing nucleoporins is required to mediate nuclear import of RanGDP. J Mol Biol 1999;293:579-93.
- Ribbeck K, Lipowsky G, Kent HM, Stewart M, Görlich D. NTF2 mediates nuclear import of Ran. EMBO J 1998;17:6587-98.
- Smith A, Brownawell A, Macara I G. Nuclear import of Ran is mediated by the transport factor NTF2. Curr Biol 1998;8:1403-6.
- Steggerda SM, Black BE, Paschal BM. Monoclonal antibodies to NTF2 inhibit nuclear protein import by preventing nuclear translocation of the GTPase Ran. Mol Biol Cell 2000;11:703-19.
- Geyer M, Assheuer R, Klebe C, et al. Conformational states of the nuclear GTP-binding protein Ran and its complexes with the exchange factor RCC1 and the effector protein RanBP1. Biochemistry 1999;38:11250-60.
- Ohtsubo M, Okazaki H, Nishimoto T. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J Cell Biol 1989;109:1389-97.
- Klebe C, Bischoff FR, Ponstingl H, Wittinghofer A. Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry 1995;34:639-47.
- Yoneda Y. Nucleocytoplasmic protein traffic and its significance to cell function. Genes Cells 2000;5:777-787.
- Moroianu J, Blobel G. Protein export from the nucleus requires the GTPase Ran and GTP hydrolysis. Proc Natl Acad Sci USA 1995;92:4318-22.
- Nemergut ME, Macara IG. Nuclear import of Ran exchange factor, RCC1, is mediated by at least two distinct mechanisms. J Cell Biol 2000;149:835-49.
- Lounsbury KM, Macara IG. Ran-binding protein 1 (RanBP1) forms a ternary complex with Ran and karyopherin β and reduces Ran GTPase-activating protein (RanGAP) inhibition by karyopherin β. J Biol Chem 1997;272:551-5.
- Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science 2002;295:2452-6.
- Takizawa CG, Weis K, Morgan DO. Ran-independent nuclear import of cyclin B1-Cdc2 by importin β. Proc Natl Acad Sci USA 1999;96:7938-43.
- Ribbeck K, Kutay U, Paraskeva E, Görlich D. The translocation of transportin-cargo complexes through nuclear pores is independent of both Ran and energy. Curr Biol 1999;9:47-50.
- Sachdev S, Bagchi S, Zhang DD, Mings AC, Hannink M. Nuclear import of IκBα is accomplished by a Ran-independent transport pathway. Mol Cell Biol 2000;20:1571-82.
- Yokoya F, Imamoto N, Tachibana T, Yoneda Y. β-catenin can be transported into the nucleus in a Ran-unassisted manner. Mol Biol Cell 1999;10:1119-31.
- Hetzer M, Mattaj IW. An ATP-dependent, Ran-independent mechanism for nuclear import of the U1A and U2B'' spliceosome proteins. J Cell Biol 2000;148:293-303.
- Faustino RS, Stronger LN, Landry MN, et al. Lysophosphatidylcholine stimulates nuclear protein import in vascular smooth muscle cells via stimulation of RanGAP activity. Mol Pharmacol 2007;71:438-45.
- Richard MN, Deniset JF, Kneesch AL, et al. Mechanical stretch stimulates smooth muscle cell growth, nuclear protein import and nuclear pore expression through mitogen-activated protein kinase activation. J Biol Chem 2007;282:23081-8.
- Czubryt MP, Austria JA, Pierce GN. Hydrogen peroxide inhibition of nuclear protein import in aortic vascular smooth muscle cells is mediated by the MAP kinase ERK2. J Cell Biol 2000;148:7-15.
- Zettler M E, Prociuk M A, Austria J A, et al. Oxidized low density lipoprotein retards the growth of proliferating cells by inhibiting nuclear translocation of cell cycle proteins. Arterioscl Thromb Vasc Biol 2004;24:1-7.
- Chahine MN, Dibrov E, Blackwood DP, et al. Oxidized LDL enhances stretch-induced smooth muscle cell proliferation through alterations in nuclear protein import. Can J Physiol Pharmacol 2012;90:1559-68.
- Chahine MN, Mioulane M, Sikkel M B, et al. Nuclear pore rearrangements and nuclear trafficking in cardiomyocytes from rat and human failing hearts. Cardiovasc Res 2015;105:31-43.
- Chahine M N, Pierce G N. Therapeutic targeting of nuclear protein import in pathological cell conditions. Pharmacol Rev 2009;61:358-72.