Virologic failure on anti-retroviral therapy without HIV drug resistance mutation
Received: 12-Feb-2018 Accepted Date: Mar 01, 2018; Published: 10-Mar-2018
Citation: Dutta N, Omesh S, Nandi S, et al. Virologic failure on anti-retroviral therapy without HIV drug resistance mutation. J Immune Disord Ther 2018;2(1):1-5.
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
BACKGROUND: HIV drug resistance (HIV DR) compromises the antiretroviral therapy (ART) outcome. This study aimed to detect presence of different HIV DR Mutations (DRM) in patients on 2nd line ART failure in Eastern India.
METHODS: HIV/AIDS patients on 2nd line ART were evaluated for virologic failure. HIV-1 genotyping was performed for the virologic failure samples employing Viroseq (Abbott Diagnostics) assay. HIVDR profile and subtype were detected using Stanford HIV sequence database following HIV DRM definition of WHO Surveillance mutation list 2009.
RESULTS: Virologic failure (HIV 1 viral load >1000 copy/ml) was detected among 15 samples out of total 365 HIV/AIDS patients (on 2nd line ART) recruited. Genotyping was successful for 9 samples having >2000 HIV/ml and remaining 6 samples could not be genotyped due to low viral copy. DR mutations were detected in 5 out of 9 samples and among the rest 4 samples no HIV DR mutation was detected. Among NRTI based drugs, M184V and M41L were the predominant mutations (80%). For NNRTI based drugs, A98G and Y181C were predominant (80%), conferring resistance to DLV and NVP. Again for PIs, l54V, A71V, V82A and M46L were seen in 40% of the cases conferring resistance to IDV, SQV, LPV, NFV and ATV.
CONCLUSION: Study on virologic failure in the absence of HIV DR mutations might help in understanding further the HIV DR dynamics as well as planning for better clinical management for patients with HIV/AIDS.
Keywords
HIV/AIDS; ART; Virologic Failure; Drug Resistance Mutation; Reverse Transcriptase Inhibitor; Protease Inhibitor
Introduction
Anti-retroviral therapy (ART) has successfully helped managing HIV/ AIDS, but lead to emergence of HIVDR mutations [1,2]. Failing ART among people living with HIV (PLHIV) increases the possibility of emergence of HIVDR [3,4]. Failing second line ART necessitates the need of third-line ART. HIV viral load (VL) assessment is crucial requirement in ART programme. World Health Organization (WHO) advocates HIV VL monitoring to identify virologic failure as an early and precise indication of ART failure. Consequent corrective procedures would reduce accumulation of drug-resistance mutations and improve clinical outcomes [5]. However, VL testing is not routinely done in many countries due to high cost and lack of facility. HIV epidemic is nearing four decades with different ART regimens. Thus, anti-retroviral (ARV) drug resistant viruses in treatment-experienced patients are also being encountered increasingly [6]. The second line ART regimens constitute zidovudine (ZDV), lamivudine (3TC), tenofovir (TDF), and boosted lopinavir/ritonavir (LPV/r). These were introduced in India in a restricted phase wise manner [7].
The criteria to shift to second line ART are failure in clinical and/ or immunological and/ or virologic parameters in a patient on first-line ART for 6 months or more. An important target of ART is Protease (PR) that is vital for HIV replication and crucial for completion of its life cycle. Different PR inhibitors (PIs) have been approved by regulatory authorities and are in use. PIs are administered as single drug with 2 nucleoside reverse-transcriptase inhibitors (NRTIs) as second-line ART. Since 2005, boosted PIs are endorsed in combination with low-dose RTV to heighten the level of companion PIs. When PI-based ART fails, viral variants resistant to PIs emerge [8-13]. Major mutations due to amino acid substitutions alter the PR catalytic activity that consequently affects virus replication capacity [14-16]. Conversely, emergence of compensatory mutations restores replication capacity [17]. Moreover, mostly the drug resistance mutations of PR also confer cross resistance to other PIs in the class. These mutations seem essentially class specific rather than drug specific [18,19]. Boosted PI based combination ART is an efficient strategy post NNTRI-based first line ART failure. PI resistance at NNRTIbased first-line ART failure is uncommon [19].
Thus, resulting PI-based second-line ART becomes highly potent [20]. Introduction of generic first-line drugs along has considerably reduced the morbidity and mortality globally. However, in resource-limited settings, HIV viral load monitoring is rarely feasible [20]. Due to absence of VL monitoring, early virologic failure remains undetected and treatment continues to be on the failing regimen until patient develops clinical or immunologic failure resulting accumulation of drug resistance mutations [21]. High levels of HIV DR to first-line ART have been reported widely [21-24]. As a result treatment has been upgraded to PI-based second-line ART regimens in India. However, the second line ART program is somewhat limited in Indian context. Without drug resistance assay and regular virological failure monitoring, the consequences of second line therapy outcomes might remain unclear also. It is therefore, essential to assess the emergence of HIV drug resistance mutations beside clinical, virological, and immunological parameter for treatment outcomes for patients switched to second line ART. This study, first of its kind from eastern India, aimed to investigate the extent HIVDR mutations among virologic failure PLHIV on second-line ART in Kolkata.
Materials and Methods
Study setting and population
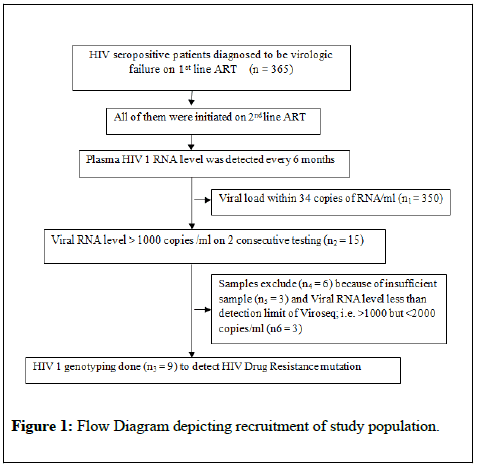
The study was conducted at Virology Laboratory, ICMR-National Institute of Cholera and Enteric Diseases, Kolkata. A total of 365 HIV/AIDS patients on 2nd line ART were recruited over a period of five years during January 2009 to December 2013. The study procedures followed Helsinki declaration 1975 and revision in 2000. The inclusion criteria were (i) consenting adult HIV/AIDS patients, age >18 years and on 2nd line antiretroviral therapy and (ii) Virologic failure (plasma HIV 1 RNA level more than 1000 copies/ml) on 2 consecutive testing on 6 month interval. Exclusion criteria were if (i) blood sample was insufficient for sequencing
(ii) missing in follow up and (iii) patients were diagnosed with HIV 2 and/or HIV 1/2 coinfection materials.
Specimens
Approximately 10 ml blood sample was collected from each participant. An aliquot of 3 ml sample was used for CD4+ T cells count estimation. Plasma was separated from remaining sample within 6 hours of collection and stored at -70°C in 1 ml aliquots. Guidelines of WHO and HIV Res Net Laboratory Working Group resistance testing were followed for HIV-1 RNA quantification (plasma VL) and genotyping for HIVDR mutation [25].
Viral load testing and CD4 estimation
Viral load assay was performed employing AMPLICOR HIV-1 Monitor Test, version 1.5 (Roche Molecular Systems Inc., Branchburg, NJ, USA). CD4/CD8+ T cell counts were estimated using (BD FACS CALIBUR, BD Biosciences, CA, USA) flow cytometer.
HIV-1 genotyping
HIV-1 genotyping was done using the ViroSeq HIV-1 Genotyping Systems (Abbott diagnostics, Wiesbaden, Germany) by sequencing the 1.8 kb protease-RT region of HIV-1 pol gene following manufacturer’s instructions. RNA was extracted from 500 μL of plasma employing guanidine-thiocynate extraction method. Reverse transcription polymerase chain reaction (RT-PCR) and then PCR was carried out for generating an amplicon of 1.3 kb. The amplicons were purified using silica spin columns. PCR products were evaluated on 1% agarose gel against 2 mass ladders for semi-quantitation of DNA. For sequencing, DNA was diluted according the band intensity found on agarose gel electrophoresis, and PCR product bands with DNA >20 nanogram were selected. Selected DNA were added to a 96-well reaction plate containing premixed Big Dye sequencing primers A, B, C, F, G, and H. Sequencing was carried out on automated (16 capillary) ABI PRISM 3100xl Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) using data collection software v3.0 and sequence analysis software v5.3. ViroSeq HIV-1 Genotyping System Software v2.8 was used to assemble the chromatographs from the seven primers and generated a contiguous sequence spanning the entire protease gene, and up to codon 335 of the reverse transcriptase (RT) gene. Points of variance were identified comparing the consensus sequence to a known reference strain, HXB-2. The sequences were edited manually and saved in FASTA format. Formatted sequences were submitted to Stanford HIV RT and Protease sequence database to determine the HIVDR mutation profile as defined in WHO Surveillance mutation list 2009 proposed by Bennett et al. and HIV-1 subtype of each sample [26,27].
Quality control
High-positive, low-positive and negative control samples were run for HIV-1 genotyping with every batch as per ViroSeq kit protocol. The positive controls were to ensure the RT-PCR and genotyping success. For ensuring good sequence quality, the high-positive control was sequenced prior to genotyping the HIV-1 clinical samples, excluding editing mistakes.
Clade typing
REGA HIV-1 subtyping tool from Stanford HIV drug-resistance database (http://hivdb.stanford.edu/) was employed to define HIV-1 subtype. The worldwide subtype reference data bases from the Los Alamos HIV database were utilized for clade typing.
Statistical analysis
Data was analysed employing Microsoft Excel using Annova for clinical and biological characters of patients to find out frequency (%) for the categorical statistical parameters.
Results
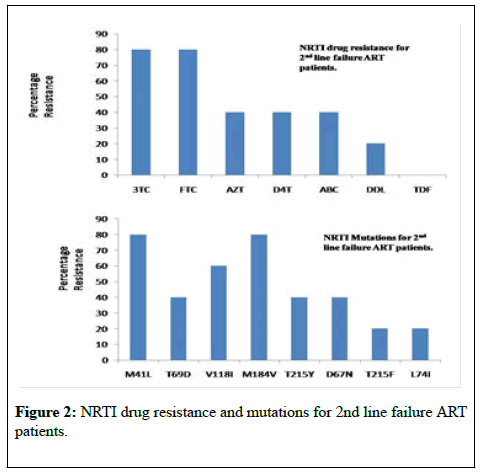
The viral loads of samples (n=9) genotyped were within the limit of 2990-4.49×106 RNA copies/ml. All HIV-1 samples were detected as subtype C (Table 1). Median age of the study group was 36 years (range: 30-46 years). The median CD4 count was 110 cells/μL (range: 51-475 cells/μL), and the median plasma HIV RNA load was 41800 copies/mL (4.60 log10), range: 2990(3.47 log10) – 4490000 (6.65 log10). HIV DRMs were detected from 5 patients, while from other 4 patients no major or minor HIVDR was detected. The Drug Resistance result showed that (for NRTI based drugs) resistance to drugs 3TC and FTC was among 80% of the patients. AZT, D4T and ABC resistance was seen in 40% of the patients. DDL resistance was seen in 20% of the patients (Figure 1 and Figure 1A). M184V that imparts resistance to NRTIs, lamivudine, and emtricitabine, was seen among 80% of the test individuals. M41L mutation was also prevalent in 80% of the cases.
| Variables | Summary |
|---|---|
| Age (yrs), median(IQR) | 36 (30-46) |
| Male | 89% |
| Female | 11% |
| Median CD4Tcell count, cells/µL (IQR) | 110(51-475) |
| Median Viral load, log10 copies/mL (IQR) | 4.60(3.47-6.65) |
| Viral load (log10 copies/mL) | |
| <4.0 | 4(44.4%) |
| 4.0–4.9 | 3(33.3%) |
| ≥5.0 | 2(22.2%) |
| HIV-1 subtypes, (%),Subtype C | 100% |
Table 1: Variables of PLHIV (n=9) failing on second-line ART genotyped.
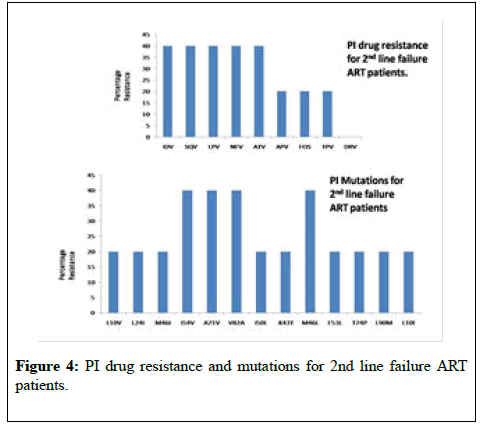
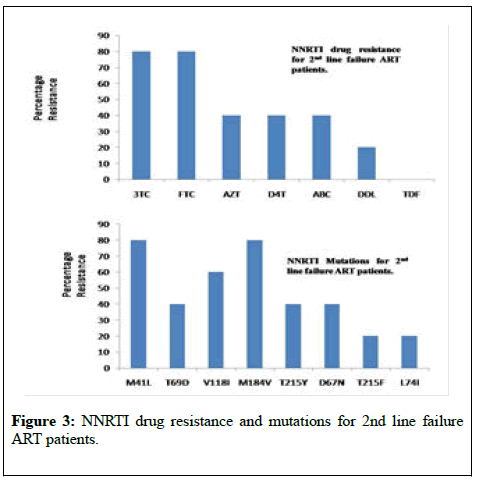
NRTI mutations T69D, T215Y, D67N were seen in 2 individuals whereas accessory NRTI mutations T215F and L74I were seen in one patient each. For NNRTI based drugs, NNRTI mutation, A98G and Y181C was predominant with 80% individuals, conferring resistance to DLV and NVP followed by K101E and G190A(40%), conferring resistance to EFV and ETR. Patients having NNRTI mutations were mostly due to the limited impact of NNRTI resistance mutations on viral fitness. A98G and Y181C were the predominant NNRTI mutations observed and these have little or no effect on replication capacity. Accessory NNRTI mutations K103N, E138K and V108I were seen in one patient each (Figure 2). PI mutations, l54V, A71V, V82A and M46L were seen in 40% of the individuals conferring resistance to drugs IDV, SQV, LPV, NFV and ATV. Accessory minor PI mutations L10V, L24I, M46I, I50L, K43T, F53L, T74P, L90M and L10I were seen in one patient each (Figure 3). Among the secondary PR mutations, amino acid variants at 7 polymorphic positions (codons 10, 20, 36, 63, 71, 77, and 93) were prevalent.
These mutations do not cause drug resistance by themselves but increase drug resistance when present together with other mutations thereby compensating the decrease in catalytic efficiency caused by other PR mutations. I54V, A71V, V82A and M46Lwere predominantly observed among participants in this study. Out of the 9 patients studied for 2nd line ART failure, in 4 PLHIVs no mutation was found with reference to Stanford HIV drug-resistance database (http://hivdb.stanford.edu/) for such sequences. One of the individuals showed only NNRTI mutations A98G, K103N, Y181C conferring resistance to DLV, NVP and EFV (Table 2: for drug name abbreviations).
| Abbreviation | Full Name |
|---|---|
| 3TC | lamivudine |
| ABC | abacavir |
| APV | amprenavir |
| ATV | atazanavir |
| ATV/c | atazanavir/cobicistat |
| ATV/r | atazanavir/ritonavir |
| d4T | stavudine |
| ddI | didanosine |
| DLV | delavirdine |
| DRV | darunavir |
| DRV/c | darunavir/cobicistat |
| DRV/r | darunavir/ritonavir |
| EFV | efavirenz |
| EFV/c/TDF/FTC | efavirenz/cobicistat/tenofovir disoproxil fumarate/emtricitabine |
| ETR | etravirine |
| FTC | emtricitabine |
| IDV | indinavir |
| LPV | lopinavir |
| LPV/r | lopinavir/ritonavir |
| MVC | maraviroc |
| NFV | nelfinavir |
| NVP | nevirapine |
| RAL | raltegravir |
| RPV | rilpivirine |
| RTV | ritonavir |
| SQV | saquinavir |
| SQV/r | saquinavir/ritonavir |
| TDF | tenofovir disoproxil fumarate |
| TPV | tipranavir |
| TPV/r | tipranavir/ritonavir |
Table 2: Drug name abbreviations
Discussion
Studies on HIV drug resistance mutations among second line ART failure patients from eastern India reveals mutations among NRTI based drugs predominantly for M184V and M41L, and for NNRTI based drugs, A98G and Y181C with 80% conferring resistance to DLV and NVP. For PI mutations, l54V, A71V, V82A and M46L were seen in 40% of the individuals conferring resistance to drugs IDV, SQV, LPV, NFV and ATV. However, the most interesting observation was absence of HIV DR mutation in 4 patients out of 9 patients on 2nd line ART failure. Pattern of mutations suggests that majority of the patients participated in this study remain susceptible to DRV/r, followed by TPV/r. Of the 2 pathways that contribute to LPV resistance the IDV-like pathway caused by mutations at positions l54V, A71V, V82A and M46L was frequently observed among the ATV- and IDV-exposed groups. This could be because patients in this study had been increasingly substituted from IDV- to ATV-based regimens. Randomized clinical trials in developed countries show that the combination of raltegravir, ETR, and DRV/r was well tolerated and was associated with the rate of virologic suppression similar to that expected in treatment-naive patient [28].
This is highly contrasting compared to 100% HIV DR mutilations observed among patients on first line ART failure from the same geographical region [29]. Adherence factors might be responsible for the study patients failing second-line ART rather than HIV strains resistant to ARV drugs, which corroborates to observation among programmatic cohorts in resource limited settings [30]. HIV drug resistance surveillance outcome among patients failing second-line ART helps in planning evidence driven treatment regimens for patients needing third-line ART. Availability of ARV drugs for those failing second-line ART might be an issue. Failing second line ART that we have encountered here necessitates planning for 3rd line ART program ensuring affordable and accessible supplies of darunavir and raltegravir.
With the introduction of ART, up to 80% decline in rates AIDS associated death has been reported [31] but it also leads to the steady rise of HIV DR mutant variants that are the major cause of ART failure [32-35]. These variants become predominant over time and pose a major challenge for both ART exposed as well ART naïve patients [36]. The gradual transmission of resistant mutants from ART-experienced patients to ART naïve individuals was reported from developed countries with even having good access to antiretroviral drugs [37-40]. However, of late, scaling-up of ART lead to the emergence of mutations in resource limited countries too [41]. WHO recommends periodic surveillance of transmitted drug resistance (TDR) mutations in ART naïve newly infected PLHIV in distinct geographical areas [42].
Currently to optimize ART regimen for scaling up, routine assessment of drug resistance-associated mutations prior to ART initiation is recommended where primary resistance has been consistently recorded [43]. Deficiency in baseline HIVDR data, interruption in treatment and improper administration of drug regimens could be the major factors of occurrence and expansion of drug resistance in resource limited countries.
Conclusion
Emergence of HIV drug resistance and virologic failure are major challenges for ART. Investigations on emergence of HIV DR mutations among patients on ART as well as virologic failure in the absence of HIV DR mutations might help in better understanding the HIV DR dynamics and planning appropriate clinical management for patients with HIV.
Acknowledgements
The authors would like to acknowledge the staffs for support in the course of this study.
Disclosure Statement
Authors have seen and approved the manuscript being submitted and there is no conflict of interest. We warrant that the article is the Authors' original work. This research has not been submitted for publication nor has it been published in whole or in part elsewhere. Authors listed on the title page have contributed significantly to the work, have read the manuscript, attest to the validity and legitimacy of the data and its interpretation, and agree to its submission to the Journal for publication
REFERENCES
- Smith DM, Schooley RT. Running with scissors: using antiretroviral therapy without monitoring viral load. Clin Infect Dis 2008;46(10):1598-600.
- Balakrishnan P, Solomon S, Kumarasamy N, et al. Low-cost monitoring of HIV infected individuals on highly active antiretroviral therapy (HAART) in developing countries. Indian J Med Res 2005;121(4):345-55.
- Vidya M, Saravanan S, Uma S, et al. Genotypic HIV type-1 drug resistance among patients with immunological failure to first-line antiretroviral therapy in south India. Antivir Ther 2009;14(7):1005-9.
- Hira SK, Panchal K, Parmar PA, et al. High resistance to antiretroviral drugs: the Indian experience. Int J STD AIDS 2004;15(3):173-7.
- Deshpande A, Jauvin V, Magnin N, et al. Resistance mutations in subtype C HIV type 1 isolates from Indian patients of Mumbai receiving NRTIs plus NNRTIs and experiencing a treatment failure: resistance to AR. AIDS Res Hum Retroviruses 2007;23(2):335-40.
- World Health Organization, WHO Res Net, including Jordan MR, Barcarolo J, Parkin N, de Oliveira T, Bertagnolio S. WHO HIV drug resistance report 2012, WHO report. Geneva, Switzerland: WHO; 2012.
- Zhang F, Dou Z, Ma Y, et al. Five year outcomes of the China National Free Antiretroviral Treatment Program. Ann Intern Med 2009;151:241-51.
- Me´decins Sans Frontie`res (MSF) (2013). Access campaign. Untangling the web of antiretroviral price reductions. 16th ed.Geneva, Switzerland: Me´decins Sans Frontie`res.
- Condra JH, Holder DJ, Schleif WA, et al. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J Virol 1996;70(12):8270-6.
- Hertogs K, Bloor S, Kemp SD, et al. Phenotypic and genotypic analysis of clinical HIV-1 isolates reveals extensive protease inhibitor cross-resistance: a survey of over 6000 samples. AIDS 2000;14:1203-10.
- Miller V. Resistance to protease inhibitors. J Acquir Immune Defic Syndr 2001;26:S34-50.
- Kandathil AJ, Kannangai R, Verghese VP, et al. Drug resistant mutations detected by genotypic drug resistance testing in patients failing therapy in clade C HIV-1 infected individuals from India. Indian J Med Microbiol 2009;27(3):231-6.
- Gupta A, Saple DG, Nadkarni G, et al. One-, two-, and three-class resistance among HIV-infected patients on antiretroviral therapy in private care clinics: Mumbai, India. AIDS Res Hum Retroviruses 2010;26:25-31.
- Orrell C, Walensky RP, Losina E, et al. HIV type-1 clade C resistance genotypes in treatment-naive patients and after first virological failure in a large community antiretroviral therapy programme. Antivir Ther 2009;14(4):523-531.
- Walmsley S, Bernstein B, King M, et al. Lopinavir-ritonavir versus nelfinavir for the initial treatment of HIV infection. N Engl J Med 2002;346(26):2039-2046.
- Chen Z, Li Y, Schock HB, et al. Three-dimensional structure of a mutant HIV-1 protease displaying cross-resistance to all protease inhibitors in clinical trials. J Biol Chem 1995;270(37):21433-6.
- Lin Y, Lin X, Hong L, et al. Effect of point mutations on the kinetics and the inhibition of human immunodeficiency virus type 1 protease: relationship to drug resistance. Biochemistry 1995;34(4):1143-52.
- Hertogs K, Van Houtte M, Larder BA. Testing for HIV-1 drug resistance: new development and clinical implications. Rec Res Dev Antimicrob Agents Chemother 1999;3:83-104.
- Kemper CA, Witt MD, Keiser PH, et al. Sequencing of protease inhibitor therapy: insights from an analysis of HIV phenotypic resistance in patients failing protease inhibitors. AIDS. 2001;15:609-15.
- Kozal M. Cross-resistance patterns among HIV protease inhibitors. AIDS Patient Care STDs. 2004;18(4):199-208.
- Desai M, Iyer G, Dikshit RK. Advances in antiretroviral drugs. Indian J Pharmacol. 2012;44:288-98.
- Radhakrishnan P. India: The Department for International Development; 2007. India and second line ART: evaluating the way forward.
- List of countries by HIV/AIDS prevalence rate 2012.
- India HIV and AIDS Statistics 2011.
- Ministry of Health and Family Welfare, Government of India 2011. NACO. National guidelines on second line ART.
- NACO. HIV Testing policy and functioning of VCTC.
- NACO. HIV Testing manual: Laboratory Diagnosis, Biosafety and Quality Control.
- Yazdanpanah Y, Fagard C, Descamps D, et al. High rate of virologic suppression with raltegravir plus etravirine and darunavir/ritonavir among treatment-experienced patients infected with multidrug-resistant HIV: results of the ANRS 139 TRIO trial. Clin Infect Dis. 2009;49(9):1441-9.
- Dutta N, Nandi S, Guha S, et al. Emergence of HIV drug-resistant mutations in East Indian population after failure of first-line antiretroviral therapy. HIV & AIDS Review. International Journal of HIV-Related Problems 2017;16(4):258-264.
- Eshleman SH, Hackett J, Swanson P, et al. Performance of the Celera Diagnostics ViroSeq HIV-1 genotyping system for sequencebased analysis of diverse human immunodeficiency virus type 1 strains. Journal of Clinical Microbiology 2004;42(6):2711-17.
- Bennett DE, Camacho RJ, Otelea D, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drugresistance. PLoS One 2009;4(3):e4724.
- Stanford University HIV Drug Resistance Database IVDR
- Bakhouch K, Oulad-Lahcen A, Bensghir R, et al. The prevalence of resistance-associated mutations to protease and reverse transcriptase inhibitors in treatment-naive (HIV1)-infected individuals in Casablanca, Morocco. J Infect Dev Ctries 2009;3(5):380-91.
- Novak RM, Chen L, MacArthur RD, et al. Prevalence of antire-troviral drug resistance mutations in chronically HIV-infected, treatment-naive patients: implications for routine resistance screening before initiation of antiretroviral therapy. Clin Infect Dis 2005;40(3):468-74.
- Re MC, Monari P, Bon I, et al. Conflicting interpretations of the prevalence of mutations associated with drug resistance in antiviral naive HIV-1 patients with acute and chronic infection. Int J Antimicrob Agents 2004;23(2):164-8.
- Re MC, Monari P, Bon I, et al. Analysis of HIV-1 drug resistant mutations by line probe assay and direct sequencing in a cohort of therapy naive HIV-1 infected Italian patients. BMC Microbiol 2001;1:30.
- Wensing AM, van de Vijver DA, Angarano G, et al. Prevalence of drug-resistant HIV-1 variants in untreated individuals in Europe: implications for clinical management. J InfectDis 2005;192(6):958-66.
- Manosuthi W, Thongyen S, Nilkamhang S, et al. HIV-1 drug resistance-associated mutations among antiretroviral-naive Thai patients with chronic HIV-1 infection. J Med Virol 2013;85(2):194-9.
- Toni T, Masquelier B, Minga A, et al. HIV-1 antiretroviral drug resistance in recently infected patients in Abidjan, Cote d'Ivoire: A 4-year survey, 2002-2006. AIDS Res Hum Retroviruses 2007;23(9):1155-60.
- Bennett DE, Bertagnolio S, Sutherland D, et.al. The World Health Organization's global strategy for prevention and assessment of HIV drug resistance. Antivir Ther 2008;13(2):1-13.
- Adolescents PoAGfAa. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents.Department of Health and Human Services 2014
- Thompson MA, Aberg JA, Hoy JF, et al. Antiretroviral treatment of adult HIV infection: 2012 recommendations of the International Antiviral Society-USA panel. JAMA. 2012;308(4):387-402.
- Vandamme AM, Camacho RJ, Ceccherini-Silberstein F, et al.European recommendations for the clinical use of HIV drug resistance testing: 2011 update. AIDS Rev 2011;13(2):77-108.