Superbug horror and its relations to antibiotics, probiotics and vitamins
Received: 03-Mar-2018 Accepted Date: Apr 27, 2018; Published: 10-May-2018
Citation: Chakraborty AK, Muneim GE, Pradhan S, et al. Superbug horror and its relations to antibiotics, probiotics and vitamins. J Pharm Toxicol. 2018;1(1):14-9.
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
Recent crisis in human health is mostly due to spread of multidrug-resistant bacteria or superbugs. Molecular analysis has indicated integrons and R-plasmids (2-15 kb) have combined with F’-conjugative plasmid (62 kb) producing large MDR-Plasmids (50-500 kb) that could donate mdr genes to all bacteria present in the intestine. We used >20,000 tons antibiotics each year to remove bacteria from our body and thus eradicated >35,000 species of gut microbiota from intestine that produce vitamins (riboflavin, pantothenate, niacin, folate, biotin, etc.). We now use probiotic capsule with ~10 bacteria (Lactobacillus, Bifidobacterium, Streptococcus, etc.) but many gut microbes are hard to grow and study in vitro. Thus, antibiotics have produced a greater crisis that human would be extinct unless mdr genes are created and diversified to withstand the new drug derivatives discovered every year! The crisis of vitamins is obvious as bacterial plasmids have been acquiring vitamin synthesizing genes and other genes related to complex biomolecules synthesis. Bacteria know that a further great war will be awaited as scientists will produce more deadly antibiotics and are acquiring multiple (20-60) gene rearrangement cassettes (IntI, Tn3, Tn5, Tn30, IS-26, etc.) in large plasmids for mdr gene creation. Moreover, intestinal cells also need lipopolysaccharides, linolinic acid, taurocholic acids and butyrate, etc., and stay in a tight symbiotic control over producing cytokines, interleukins and immuloglobins. Superbugs NDM1 Escherichia coli, MRSA Staphylococcus aureus, MDR Acinetobacter baumannii and XDR Mycobacterium tuberculosis, etc., are hard to treat due to accumulations of amp, blaNDM1, blaOXA23, tetA/C, mexAB, arr3, catB3, sul1/2, dhfr, penA, acrAB, mtrCD, aacC1, aadA2, aphA4, aacA4, mcr-1, etc., MDR genes and DEGs (acrAB, tetA/C, mcr, mtrCD, mexAB-EF) in plasmids and chromosome(s) as well as mutations in the antibiotic targets like gyrB, parC, porB, 16S rRNA, rpsL genes. WHO has warned for reduced antibiotic use and G-20 Nations are together to address the anticipated antibiotic horror, not superbug horror. Perhaps, phage therapy, gene medicines, heterogeneous phyto-antibiotics and drug nano-carriers will be future agenda to replace antibiotics.
Keywords
MDR-plasmids; Gut microbiota; Antibiotic void; Probiotics; Vitamin genes
Penicillin antibiotics discovered in 1928 by Nobel Laureate Alexander Flaming and thereafter by Dr. Selman Waksman discovered over twenty antibiotics including streptomycin [1]. In truth, sulphonamides was introduced in 1930, sulfa-drugs in 1940, penicillins in 1943, tetracycline in 1945-1951, streptomycin and chloramphenicol in 1949, erythromycin in 1952 and ciprofloxacin in 1965 [2,3]. Sadly, first report on drug resistant bacteria appeared as early as 1940 after ten years of antibiotic discovery and 3 years early of first commercialized production of Benzyl penicillin in 1943. Plasmid pBR322 was made first using restriction enzymes and DNA ligase enzymes from three natural R-plasmids followed by bacterial transformation and plasmid isolation; and we know the sequences of first mdr gene (amp) in 1965 [3]. Question arises, are rapid expansion of recombinant plasmid work with amp gene and there after neo, hph, pac, geo, aac and cat genes are producing MDR bacteria even rec- strain of Escherichia coli bacteria used in RDT work? [4,5] Anyway, whole world are safe after World War II taking antibiotics in any bacterial infections and reputed Pharmaceutical Companies have generated with great capital and share market. However, in 1960 MIC of drugs were start increasing and to keep the capital market safe, drug companies were also produced 100 new derivatives of old drugs like ampicillin, oxacillin, methicillin, cefotaxime, ceftriaxone and many strong carbapenems (imipenem, dorripenem, meropenem) that target bacterial peptidoglycan biosynthesis [5,6]. Streptomycin and tetracycline resistance appeared between 1958-1962 followed by aminoglycosides resistance in 1980 [7]. Ciprofloxacin resistance became prominent in 1985 and azithromycin resistance was vibrant between 1995-1999 [8]. Most horror reported in 2009 that blaNDM1 enzyme cleaves all penicillins, cephalosporins and carbapenems and lastly Mcr-1 enzyme was reported in 2016 that make resistant to colistin drug (an old 1945 toxic drug) by transferring phosphoethanolamine to lipid A of bacterial membrane [9]. A great horror in Pharmaceutical Industry and many companies are sold or combined to accelerate the research on novel therapeutics like gene medicines, nanocarriers, phage therapy and enzybiotics [10,11]. India uses 8 billion unit antibiotics in 2001 to 12.9 billion units in 2010 with overall 36% increase in 10 years worldwide. This suggests that it is a habit for Indian peoples to take >10 antibiotic pill per year even the infection is viral one. Sadly US peoples take average >20 antibiotic pill per year. This led to conclusion that gut is the main source of mdr genes creation and multi-resistance [11,12]. At least few millions peoples (mostly neonatal and elderly in the poor countries) are infected every month and expected 10 million death every year where potent drugs like ampicillin, tetracycline, chloramphenicol, azithromycin, streptomycin, ciprofloxacin, refampicin, sulfamethoxazole, etc., would not able to clear most infections like Salmonella typhi, Mycobacterium tuberculosis, Siegella sonni, Escherichia coli, Acinetobacter baumannii and Staphylococcus aureus [13,14]. But meropenem, ceftriaxone, lomofloxacin, linezolid, amikacin, colistin type new drug derivatives are sometime effective if the infections are not PAN drug resistant [15]. It has been accepted by WHO that antibiotics have produced ugly effects on bacteria as well as human intestinal cells, those are in tight symbiosis to produce vitamins needed for >30000 enzymatic reactions called human metabolosome [16]. Over use of antibiotics indeed has killed vital gut microbiota (since 1943) which are now compensated by taking multivitamins (B1, B2, B6, B12, A, C, D) and probiotic capsules [17]. We have discussed here the anticipated side effects of antibiotic uses that have killed useful gut bacteria producing mdr genes horror of drug industry and human as well as animal health.
Materials and Methods
Water from Ganga River was collected at the morning from Babu Ghat (Kolkata-700001) and Howrah Station River [16]. About 100 μl of water was spread onto 1.5% Luria Bartoni-agar plate containing different concentration of antibiotics at 2-50 μg/ml. MDR bacteria were selected in agar-plate containing ampicillin, streptomycin, chloramphenicol, tetracycline or ciprofloxacin at 50, 50, 34, 20 μg/ml, respectively. As imipenem and meropenem resistant bacteria were present low (0.08-0.2 cfu/ml water), a modified method was followed. 2 ml 5x LB media was added into 10 ml River/Sea water at 2-10 μg/ml concentration and was incubated 24 h to get drug resistant bacteria population. Meropenem resistant bacteria were further selected on tetracycline, chloramphenicol and streptomycin to get the superbugs. Antibiotics were purchased from HiMedia and stored at 2-50 mg/ml at -20°C [18,19]. Antibiotic papers were also purchased from HiMedia according to CLSI standard. Antibiotic papers are: A-25 (ampicillin), T-10 (tetracycline), AT-50 (aztreonam), COT-25 μg (Cotrimoxazole), Met-10 μg (methicillin), CAZ-30 μg (ceftazidime), LOM-15 μg (lomofloxacin), VA-10 μg (vancomycin), AK-10 μg (Amikacin), TGC-15 μg (tigecycline), LZ-10 μg (linezolid), and IMP-2 μg (imipenem). The plasmid DNA was isolated from overnight culture using Alkaline-Lysis Method [20,21]. 16S rDNA gene colour Sanger’s di-deoxy sequencing was performed by SciGenom Limited, Kerala, India). PCR amplification was performed using 1 unit Taq DNA polymerase, 20 ng DNA template, 0.25 mM dXTPs, 1.5 mM MgCl2, for 35 cycles at 95°C/30” (denaturation)-52°C/50”(annealing)-72°C/1.5’ [22]. The product was resolved on a 1% agarose gel in 1x TAE buffer at 50 V for 2-4 h and visualized under UV light and photograph was taken. The primers for 16S rRNA amplification and mdr genes are given below. NCBI BLAST analysis was performed for bacterial specific gene analysis (http://www.ncbi. nlm.nih.gov/blast) and data was submitted to GenBank. NCBI databases were retrieved using the BLAST programmes [21-23]. The complete genes are sequenced in plasmids and were analyzed by Seq-2 programme of BLAST [24]. Multalin protein sequence software was used to get the nature of conserved sequences among metallo-class B β-lactamases [23]. Sometime, diverged sequences are manually cut and paste into align position in MS word so that it is appeared both sequences have similarity. For retrieving any nucleotide, we type the same at the NCBI port (http://www.ncbi.nlm.nih.gov/ nucleotide or Protein] and to BLAST search to type the accession number for protein or DNA into BLAST port (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch] [25-27].
Results and Discussion
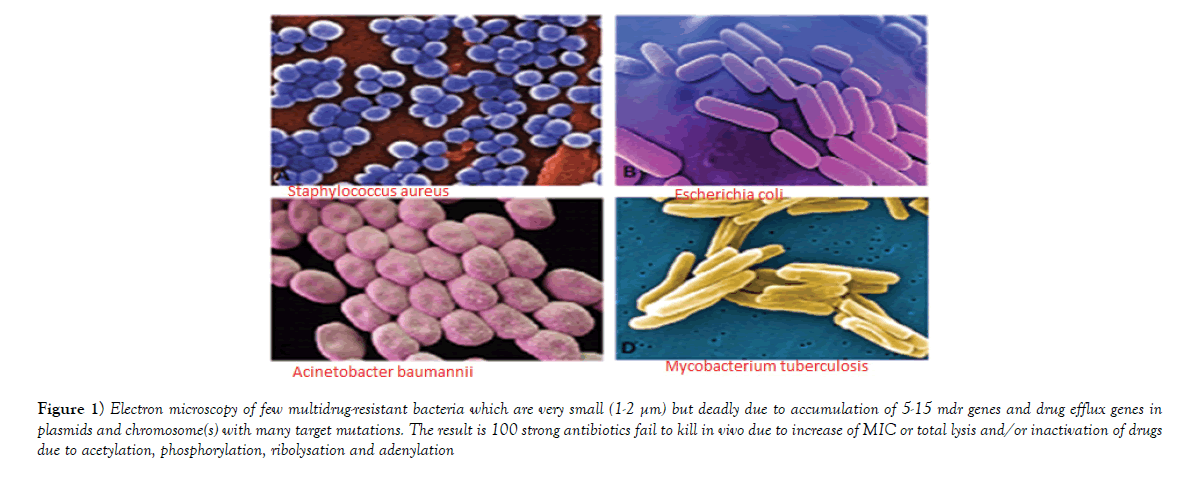
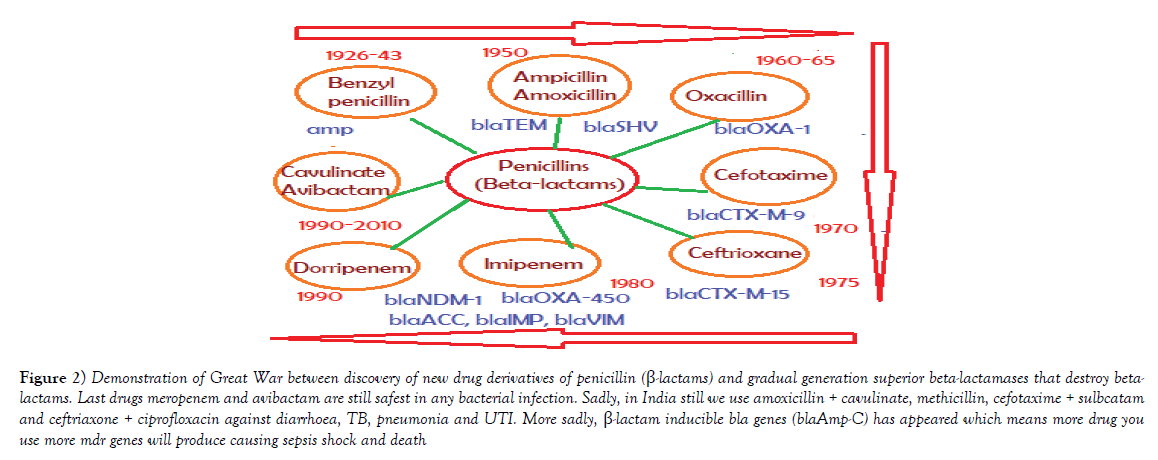
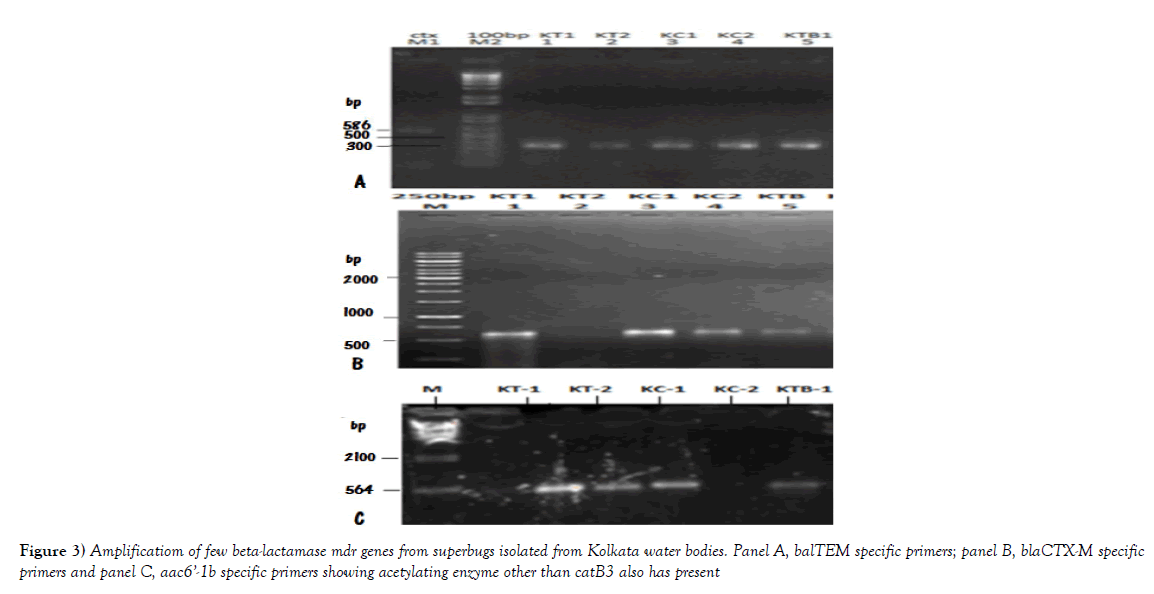
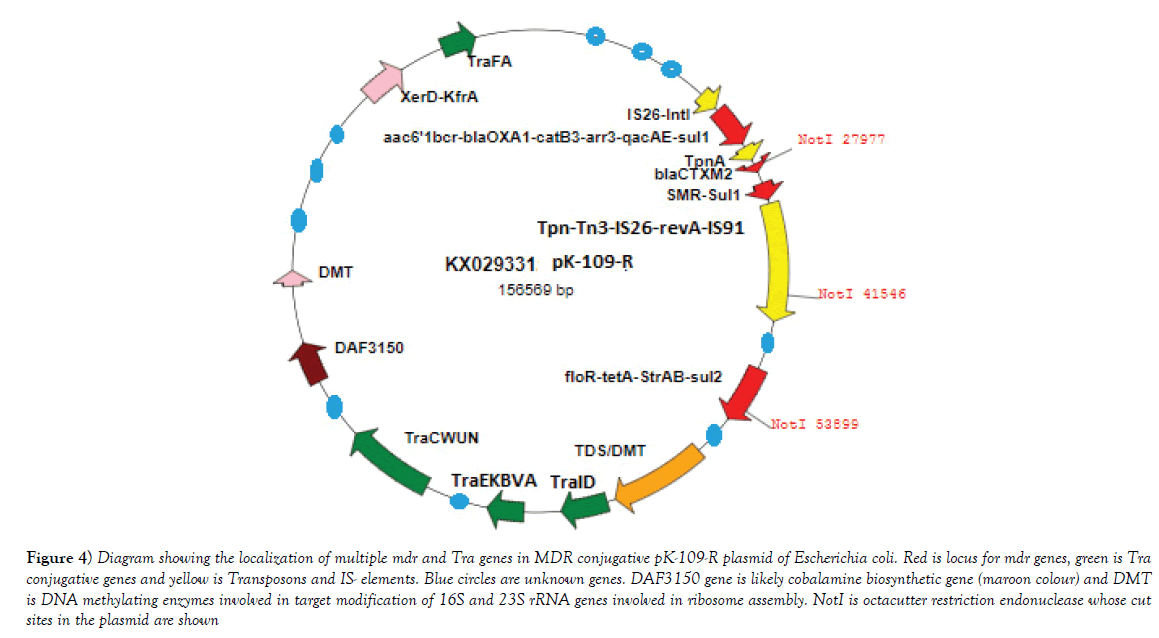
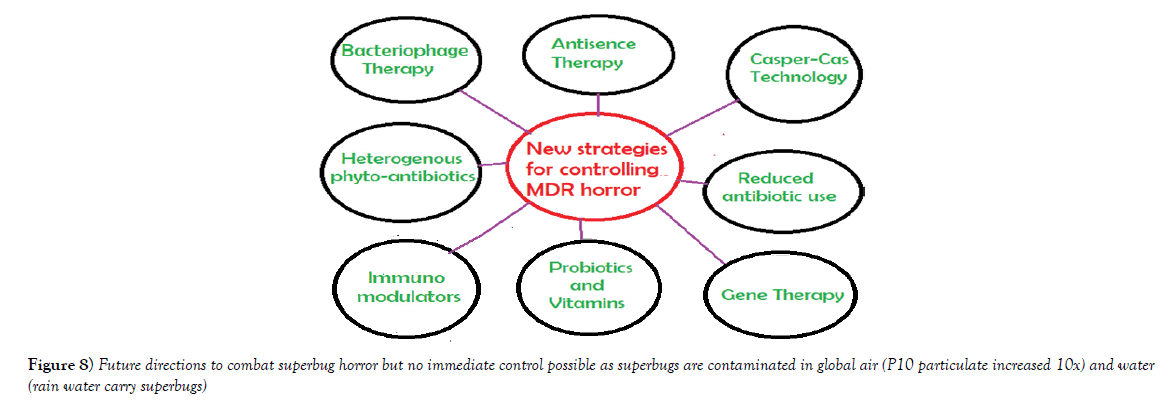
Multidrug-resistant bacteria were contaminated in world water as PM10 particulate (200-400 μg/m3) in air carry bacterial spores and during rain drop anywhere. Table 1 shows the recently isolated plasmids from MDR pathogens carrying multiple mdr genes (amp, blaNDM1, blaOXA, tet, mexAB, arr3, cat, sul, dhfr, penA, acrAB, aacC1, aadA2, aphA4, aacA4, mcr-1). Indeed such GenBank search indicated a rapid acquisition of multiple mdr genes in Escherichia coli, Mycobacterium tuberculosis, Acinetobacter baumannii and Salmonella typhi, etc., is shocking and now >35000 species gut microbiota were antibiotic resistant Figure 1 for EM structures of 1-2 μm superbugs). The antibiotics were available in 1940s onwards for all and we ingested at least 20 great antibiotics (ampicillin, tetracycline, chloramphenicol, erythromycin, oxacillin, sulfamethoxazole, streptomycin, etc.) between 1950-1960. Benzyl penicillin was discovered in 1928 and presence of penicillin resistant bacteria was detected in 1940 but during World War I and World War II Benzyl penicillin was only available to soldiers to stop infections of injuries [27]. (Figure 2) shows how new derivatives of penicillin were made following the genesis of new beta-lactamases that cleave beta-lactam ring and inactivate the drug. Drug industry is always run to discover new derivatives of penicillin, the wonder drug that stop bacterial cell wall peptidoglycan biosynthesis and we have no cell wall being non-toxic [27]. Scientists made cefalosporins changing 5-membered B-ring into 6-membered ring (cefoxitin, cefotaxime and ceftrioxane). But blaCTX-M enzymes were generated soon and now very prominent in many MDR plasmids. Then new carbapenem derivatives were introduced by changing S-atom in B-ring into C-atom (imipenem, ertapenem, dorripenem, meropenem). But blaVIM, blaIMP, blaNDM1 enzymes were appeared in plasmids that inactivate not only ampicillin but cefotaxime as well as imipenem. (Figure 3) shows the amplification of blaTEM (panel A) and blaCTX-M (panel B) genes from isolated plasmids of superbugs that resistant to many drugs (ampicillin, tetracycline, cefotaxime, azithromycin) [18]. The situation is so frustrating that drug companies merged for more basic research as drug development needs >2 billion dollars and ten years research for clinical study. Tetracycline drug was useless due to tetC efflux protein and tetM protein that has strong affinity for tetracycline. Streptomycin was inactivated by strA and strB enzymes that phosphorylate and inactivate streptomycin. Similarly, acetylating enzymes (CAT, AAC) inactivate chloramphenicol and aminoglycoside antibiotics as well as ciprofloxacin and those drugs usually prevent protein synthesis in the ribosome [10]. Figure 3, panel C shows the presence of AAC6’-1b genes in multidrug-resistant bacteria isolated in Kolkata water bodies. Moreover, mutations in the gyrAB and parC protect MDR bacteria against fluroquinolones (norfloxacin, lomofloxacin). Arr3 gene ribosylates rifampicin and inactivate it preventing to act on bacterial RNA polymerase. Figure 4 shows the structure of a conjugative MDR plasmid (pK- 109-R) with many mdr genes activated by many transposons and many Tra genes. We are yet unable to fully sequence large plasmids of our isolates from India. Such study clearly indicated that MDR bacteria are abundant as have described worldwide with activation of mdr genes and probably other genes involved in vitamin synthesis [28]. Figure 5 shows the activation of pigment production in Pseudomonas aeruginosa DB-1_mdr (accession no.KY769875) induced by Beta-lactam drug (lane 3). Such study is important as WHO has reported the escalated unnecessary of antibiotic use worse the condition of patients increasing drug MIC due to over expression of Beta-lactamases (27). We previously showed that acrAB, mcr and tetC drug transporters were activated in Kolkata superbugs [18,29,30]. The analysis showed a escalated antibiotic use has killed gut microbiota that synthesized 20 vitamins and biomolecules needed for >30,000 enzymatic reactions favouring normal human metabolosome (glycolysis, TCA cycle, electron transport in the mitochondrial membrane and metabolism of fat, protein and nucleic acids). We have raised the strong voice that we must use multi-vitamin complex after each antibiotic dose and for regeneration of the gut bacteria, we must use probiotic (bifidobacterium) capsule. Sadly, many gut bacteria are hard to grow in vitro as live in a tight symbiotic relation with intestinal cells that secrete interleukins and cytokines. High quality research from US Human Microbiome Project (HMP), European Metagenomics of the Human Intestinal Tract (MetaHIT) and others have demonstrated the beneficial functions of the normal gut flora (>35000 species) on health. Figure 6 shows a complex relation between gut microbiota and intestinal luminal cells [30]. So repeated dose of antibiotics are not good and vigilance of probiotic use must be increased as I am advised amoxicillin+cavulinate but not vitamins or probiotics are prescribed recently. The situation in poor countries (Africa, Latin America and Asia) is more severe. Thus we have presented a relation among superbugs, antibiotics, probiotics and vitamins in Figure 7. We cannot ignore any one of the four parameters as (i) 95% clinical bacteria are now ampicillin drug resistant, (ii) critical messages have generated between bacteria and intestinal cells to make more mdr genes in plasmids, (iii) mdr genes have moved into bacterial chromosome, (iv) many integrons, transposons and IS-elements assembled in MDR plasmids, (v) gut bacteria research has got momentum from US Human Microbiome Project (HMP) and European Metagenomics of the Human Intestinal Tract (MetaHIT) and vitamins are available and cost effective. Finally, Pharmaceutical companies are well ahead in new drug development and perhaps new medical technological advances are in the door as depicted in Figure 8. So phage therapy in centre stage [31]. New antibiotics against cancer, diabetes and mental disorders have discovered but none against infectious bacterial pathogenesis. So we hope gene therapeutics will be advanced like ribozyme therapy, Casper-Case technology [32,33] and toxic drugs will be delivered by nano-carriers like fullerenes, DNA cages and asiloglycopeptides [34]. We have MDR horror now but we will overcome the antibiotic Dark Age as Nobel Laureates Alexander Flaming and Selman Waksman did in 1928 and onwards.
Figure 1: Electron microscopy of few multidrug-resistant bacteria which are very small (1-2 µm) but deadly due to accumulation of 5-15 mdr genes and drug efflux genes in plasmids and chromosome(s) with many target mutations. The result is 100 strong antibiotics fail to kill in vivo due to increase of MIC or total lysis and/or inactivation of drugs due to acetylation, phosphorylation, ribolysation and adenylation.
Figure 2: Demonstration of Great War between discovery of new drug derivatives of penicillin (ß-lactams) and gradual generation superior beta-lactamases that destroy betalactams. Last drugs meropenem and avibactam are still safest in any bacterial infection. Sadly, in India still we use amoxicillin + cavulinate, methicillin, cefotaxime + sulbcatam and ceftriaxone + ciprofloxacin against diarrhoea, TB, pneumonia and UTI. More sadly, ß-lactam inducible bla genes (blaAmp-C) has appeared which means more drug you use more mdr genes will produce causing sepsis shock and death.
Figure 4: Diagram showing the localization of multiple mdr and Tra genes in MDR conjugative pK-109-R plasmid of Escherichia coli. Red is locus for mdr genes, green is Tra conjugative genes and yellow is Transposons and IS- elements. Blue circles are unknown genes. DAF3150 gene is likely cobalamine biosynthetic gene (maroon colour) and DMT is DNA methylating enzymes involved in target modification of 16S and 23S rRNA genes involved in ribosome assembly. NotI is octacutter restriction endonuclease whose cut sites in the plasmid are shown.
Figure 5: Growth of Pseudomonas aeruginosa DB-1_MDR (accession no. KY769875) pigment producing MDR bacteria (selected with 100 µg/ml ampicillin+ 50 µg/ml faropenem+25 µg/ml cefotaxime+5 µg/ml imipenem). Lane 1= only LB media (blank), 2= +DB-1 bacteria (-drug), 3= +Ampicillin, 4= +Feropenem, 5= +Cefotaxime and 6= +Imipenem. The dye production is facilitated with ampicillin better, than feropenem (which is penicillin type but quinolidone substituents and less affected by ß-lactamases (ESBL) but very less with cephalosporins like cefotaxime (thiazoline B ring of ß-lactam is six-member ring) and also with carbapenems like imipenem (thiazoline B ring of ß-lactam has C atom in place of S atom).
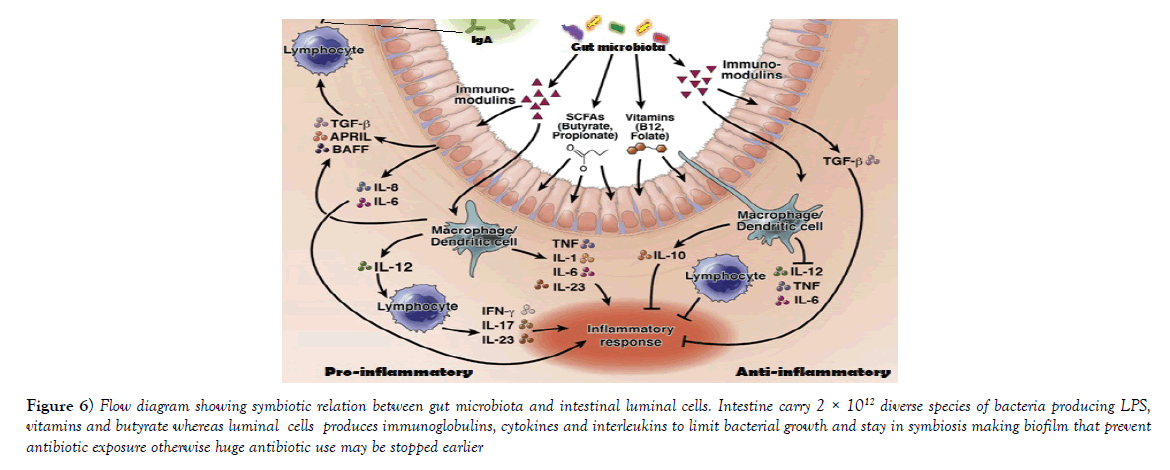
Figure 6: Flow diagram showing symbiotic relation between gut microbiota and intestinal luminal cells. Intestine carry 2 × 1012 diverse species of bacteria producing LPS, vitamins and butyrate whereas luminal cells produces immunoglobulins, cytokines and interleukins to limit bacterial growth and stay in symbiosis making biofilm that prevent antibiotic exposure otherwise huge antibiotic use may be stopped earlier.
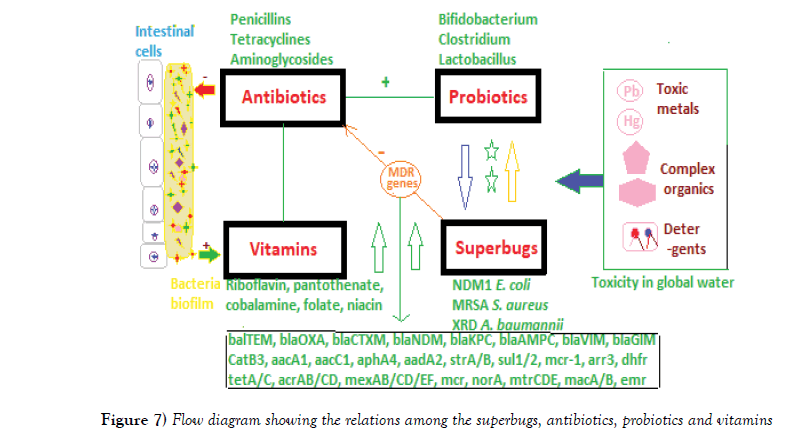
Figure 7: Flow diagram showing the relations among the superbugs, antibiotics, probiotics and vitamins.
Conclusion
Diversified MDR genes are created highly in bacterial plasmids and it appears most bacteria have received transferable small, medium and or large plasmids with 5-15 mdr genes and 10-60 transposons and IS-elements [35]. WHO and G-20 Nations recently gather in Germany for making One Nation platform and united research programmes. Indian NAP-AMR (The National Action Plan on Antimicrobial Resistance 2017-2021) has pinpointed five main areas to curve superbug horror: (i) improving awareness and understanding of AMR through effective education and surveillance, (ii) reducing infection by increasing preventive measures, (iv) reducing the use of antimicrobials in health, food animals and agriculture (v) promoting for AMR research and drug innovations and (v) strengthening India’s leadership on AMR and International collaboration (assessed on October, 2017). Practically, we have frightened to see the MDR plasmid’s size and genes are incorporated and reported a Vibrio cholera strain 2012EL-2176 harvoring IncA/C2 plasmid containing blaCMY-2, blaCTX-M-2, blaTEM-1, floR, aac(3)-IIa , strA/B, sul1/2, dfrA1, dfrA27, tetA, mphA, mdr-genes and also resistant to ciprofloxacin due to mutation in gyrA (S83I)/parC (S85L) has also seen in plasmid pMRV150 (accession no. EU116442). Bacillus thuringiensis plasmid pBMB293 (Accession no. CP007615, 294 kb) has no mdr gene but genes for enterotoxins (protein id. AIM34697), dipterans toxin (protein id. AIM34741) and reverse transcriptase, DNA polymerase β, DNA topoisomerase III and type II secretion system. Similarly, Bacillus anthrus plasmid pX01 (accession no. CM002399; 171 kb) has toxin gene (protein id. AFL55645, 809 aa) and also in pBMB293 plasmid. More and more genes in plasmids will be evident and likely phage therapy comes of age (36-38) (Table 1). Small non-coding RNAs (miRNA) have potential to therapeutic applications as in cancer but its role in MDR bacteria remains elusive (39-41). Many unknown small proteins are located in such plasmids and all have indicated alarming signal. In fact, our work has not completed yet and we argue that others work on MDR bacterial mechanism of pathogenesis is also premature. So much diversities among the mdr genes in single MDR plasmid, then by studying 2-3 genes in clinical samples is nothing conclusive [44]! The MDR plasmids have 20-50 unknown genes and we first to show that such genes actually vitamin synthesizing genes, DAF domain proteins and proteins involved in complex biomolecules synthesis [28]. We anticipate GenBank data submission should be given priority check-up and all original colour data should be submitted. We are annoyed to identify many genes have similar names but no sequence similarities at all. We can give hundred examples but unnecessary. We have to minimize research cost and also our mission will be truthful and fruitful research to reduce the drug cost! In India, we now know antibiotic is dangerous but do not know repeated doses of antibiotics how could be dangerous without taking vitamins and probiotics. Mostly antibiotic drug sensitivity tests of blood, urine and stool must mandatory before taking oral antibiotics. This is first paper that questioned many problems of modern MDR research and quick publication. We do understand that funding sources are not enough but G-20 Nations agreed in common research agenda. Our research first has shown that contamination of this Earth with MDR bacteria occurs during rain [17]. We believe this article will promote awareness on diseases as neonatal and elderly should not bath in any raw water (river, sea and pond). We hope this paper and other related papers [10] and reviews [45] will change the concept of diagnosis, treatment options and quality health in the era of superbug horror and antibiotic horror. To reduce the expenditure, one nation research programmes will be adopted and all medical discoveries must be patented by United Nations or WHO.
| pS121-1a | CP022170 | 192 kb | mexC, aac6’-1b, cmlA5, ANT3”-1a, dhfr, aac6’-1b-cr, OXA1, sul1, mphE 2’, floR | Aeromonas sulmonicida |
| pEGY1mcr-1 | CP023143 | 229 kb | Aac3’-IIa, EmaA, TelA/B/F, tetA, mcr-1, ANT3”-Ia, cmlA, sul1, floR | Escherichia coli |
| P1068-KPC | MF168402 | 146 kb | CTX-M-65, TEM-1, SHV-12, KPC-2, MerA/C, RmtB | Klebsiella pneumonia |
| PSHE-CTX-M | CP022359 | 193 kb | CTX-M-15, aac3’-IIa, CusA, tetA, aph3”-1d, aph3”-1b, sul1 (Vit*) | Shewanella bicestrii |
| pECAZ155_KPC | CP019001 | 272 kb | KPC, mphA, sul1, aac6’-II, cmlA, aph6’-1d, CMY-2, MFS, tetB, aac6’-1a | Escherichia coli |
| pKP13f | CP004000 | 295 kb | Sul1, QacE, TEM-1, OXA-9, aad3”, sul2, CTX-M-2, aac6’-Ia, TerX/X/A | Klebsiella pneumoniae |
| pAUSMDU8141 | CP022697 | 149 kb | MFS, sul1, catA2, tetD,TEM-1, aac3’-Ia, | Citrobacter farmeri |
| pKN-LS6 | NC_021654 | 246 kb | catA1, dhfr, sul1, mphA, MFS, ABC, silP/E/A/B, arsH/A, cusB/C/S | Klebsiella pneumoniae |
| pRJ119-NDM1 | KX636095 | 335 kb | Ble, TEM, cobS, aac6’, sul1, cusA,arsD, dhfr, qnr, tetA, NDM1, aph3’, sul,arsH | Klebsiella pneumoniae |
| pOZ176 | KC543497 | 501 kb | Ter2, OXA-10, MFS, TEM-8, ble, catB3, aac6’. IMP-9, neo | Pseudomonas aeruginosa |
| pKPx-2 | AP012056 | 141 kb | Aac3’/6’, catB4, tetA, sul2, OXA-1, CTX-M, TEM-1, strAB, qnrB | Klebsiella kneumoniae |
| pHX40908 | KM877269 | 249 kb | Aad, floR, hph, aac6’/3’, OXA-1, catB3, arr3, sul1, cmlA | Salmonella enterica |
Table 1: Generation of multidrug resistant plasmids with diversified mdr genes in superbugs
Acknowledgement
We thank Prof. Amiya Panda, Department of Chemistry and Chemical Technology, Vidyasagar University and Prof. Maitrayee DasGupta, Department of Biochemistry, Calcutta University, for valuable help during the study.
REFERENCES
- Yocum RR, Rasmussen JR, Strominger JL. The mechanism of action of penicillin. J Biol Chem. 1980;255:3977-86.
- Blackwood RK. Structure determination and total synthesis of tetracyclines. In: Hlavka JJ, Boothe JH, editors. Handbook of experimental pharmacology. 78, Berlin, Germany: Springer-Verlag KG. 1985;59-136.
- Finch RG. Tetracyclines. O’Grady F, Lambert HP, Finch RG, Greenwood D, eds. Antibiotic and chemotherapy. 7th ed. New York, Churchill Livingstone Ltd. 1997;469-84.
- Chakraborty AK. Multi-drug resistant genes in bacteria and 21st Century problems associated with antibiotic therapy. Biotechnol Ind J. 2016;12:114.
- Chakraborty AK. Multi-drug resistant bacteria from Kolkata Ganga River with heterogeneous mdr genes have four hallmarks of cancer cells but could be controlled by organic phyto-extracts. Biochem Biotechnol Res. 2017;5:11-23.
- Chakraborty AK. Multi-drug resistant genes in bacteria and 21st Century problems associated with antibiotic therapy. Biotechnol Ind J. 2016;12:114.
- Brown S, Amyes SGB. The sequences of seven class D ß-lactamases isolated from carbapenem-resistant Acinetobacter baumannii from four continents. Clin Microbiol Infect. 2005;11:326-9.
- Nikaido H. Multiple antibiotic resistance and efflux. Curr Opin Microbiol. 1998;1:516-23.
- Di Pilato V, Arena F, Tascini C, et al. MCR-1.2: A new MCR variant encoded by a transferable plasmid from a colistin-resistant KPC carbapenemase-producing Klebsiella pneumoniae of sequence type 512. Antimicrob Agents Chemother. 2016;60:5612-5.
- Chakraborty AK. Multi-drug resistant genes in bacteria and 21st century problems associated with antibiotic therapy. Biotechnol Ind J. 2016;12:114.
- Chakraborty AK. MDR genes are created and transmitted in plasmids and chromosomes to keep normal intestinal microbiota alive against high dose antibiotics - A hypothesis. J Mol Med Clin Appl. 2017;2:109.
- Chakraborty AK. Enzybiotics, a new class of antimicrobials targeted against multidrug-resistant superbugs. Nov Appo Drug Des Dev. 2017;2:555592.
- Fosberg KJ, Reyes A, Wang B, et al. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337:1107-11.
- Jensen KC, Hair BB, Wienclaw TM et al. Isolation and host range of bacteriophage with lytic activity against methicillin-resistant Staphylococcus aureus and potential use as a fomite decontaminant. PLoS One. 2015;10:e0131714.
- Ceccarellia D, Van Essen-Zandbergena A, Veldmana KT, et al. Chromosome-based blaOXA-48-like variants in Shewanella species isolates from food-producing animals, fish and the aquatic environment. Antimicrob Agents Chemother. 2017;61(2).
- Chakraborty, AK. Colistin drug resistant determinant Mcr-1 gene spreads in conjugative plasmids creating huge confusion for the treatment of multi-drug resistant infections. Am Res J Biotechnol. 2017;1:1-9.
- D’Costa VM, King CE, Kalan L, et al. Antibiotic resistance is ancient. Nature. 2011;477:457-61.
- Chakraborty AK. High mode contamination of multi-drug resistant bacteria in Kolkata: Mechanism of gene activation and remedy by heterogeneous phyto-antibiotics. Ind J Biotechnol. 2015;14:149-59.
- Chakraborty AK, Hodgson CP. Role of far upstream repressor elements controlling the proto-Ha-ras gene transcription. Biochem Biophys Res Commun. 1998;252:716-22.
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press, New York. 1989.
- Chakraborty AK, Das SK. Molecular cloning and characterization of the guinea pig cholinephosphotransferase gene. Biochem Biophys Res Commun. 2003;312:1104-10.
- Chakraborty AK, Zink MA, Boman BM, et al. Synthetic retrotransposon vectors for gene therapy. FASEB J. 1993;7:971-7.
- Johnson M, Zaretskaya I, Raytselis Y, et al. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008;36:W5-W9.
- Sanger, F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463-7.
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45-8.
- Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389-402.
- Chakraborty AK. Complexity, heterogeneity, 3-D structures and transcriptional activation of multi-drug resistant clinically relevant bacterial beta-lactamases. Trends Biotechnol Open Access. 2016;2:1.
- Chakraborty AK. Roles of vitamin metabolizing genes in multidrug-resistant plasmids of superbugs. BioRxiv. 2018;20.
- Delmar JA, Su CC, Yu EW. Bacterial multidrug efflux transporters. Annu Rev Biophys. 2014;43:93-117.
- Chakraborty AK. Mechanisms of AMR: Bacteria won the battle against antibiotics. Insights Biomed. 2017;2:19.
- Dalmasso M, Strain R, Neve H, et al. Three new Escherichia coli phages from the human gut show promising potential for phage therapy. PLoS One. 2016;11:e0156773.
- Hermoso JA. Taking aim on bacterial pathogens: From phage therapy to enzybiotics. Curr Opin Microbiol. 2007;10:461-72.
- Dunbar CE, High KA, Joung JK, et al. Gene therapy comes of age. Science. 2018;359(6372):pii: eaan4672.
- Chakraborty AK. Multi-drug resistant genes in bacteria and 21st century problems associated with antibiotic therapy. Biotechnol Ind J. 2016;12:114.
- McArthur AG, Waglechner N, Nizam F, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemther. 2013;57:3348-57.
- Foster SJ. Molecular characterization and functional analysis of the major autolysin of Staphylococcus aureus 8325/4. J Bacteriol. 1995;177:5723-5.
- Finch RG. Tetracyclines. Antibiotic and chemotherapy O’Grady F, Lambert HP, Finch RG, Greenwood D, eds. 7th edn. New York, Churchill Livingstone Ltd. 1997;469-84.
- RodrÃÂguez-Rubio L, Gutiérrez D, Donovan DM, et al. Phage lytic proteins: Biotechnological applications beyond clinical antimicrobials. Crit Rev Biotechnol. 2016;36:542-52.
- Yang H, Li J, Shin HD, et al. Molecular engineering of industrial enzymes: Recent advances and future prospects. Appl Microbiol Biotechnol. 2014; 98:23-9.
- Schiroli G, Genovese P, Capo V et al. Targeted genome editing in mouse hematopoietic stem/progenitor cells (HSPC) to model gene correction of SCID-X1. Hum Gene Ther. 2015;26:A8-8.
- Slaby O, Laga R, Sedlacek O. Therapeutic targeting of non-coding RNAs in cancer. Biochem J. 2017;474:4219-51.
- Timpson NJ, Greenwood CMT, Soranzo N, et al. Genetic architecture: The shape of the genetic contribution to human traits and disease. Nat Rev Genet. 2017;19: 110-24.
- Daglia M. Polyphenols as antimicrobial agents. Curr. Opin Biotechnol. 2011;23:174-81.
- Chakraborty AK. In silico analysis of hotspot mutations in the bacterial NDM-1 and KPC-1 carbapenemases that cause severe MDR phenotypes. Biochem Biotechnol Res. 2016;4:17-26.
- Chakraborty AK, Roy T, Mondal S. Development of DNA nanotechnology and uses in molecular medicine and biology. Insights Biomed. 2016;1:13.